Modules¶
- type
Pointers to arrays with accelerations, which can later be used for derived diagnostics, like energy balances.
Public Members
-
real, dimension(:,:,:), pointer mom_variables::accel_diag_ptrs::diffu => NULL() Zonal acceleration due to along isopycnal viscosity [m s-1 T-1 ~> m s-2].
-
real, dimension(:,:,:), pointer mom_variables::accel_diag_ptrs::diffv => NULL() Meridional acceleration due to along isopycnal viscosity [m s-1 T-1 ~> m s-2].
-
real, dimension(:,:,:), pointer mom_variables::accel_diag_ptrs::cau => NULL() Zonal Coriolis and momentum advection accelerations [m s-2].
-
real, dimension(:,:,:), pointer mom_variables::accel_diag_ptrs::cav => NULL() Meridional Coriolis and momentum advection accelerations [m s-2].
-
real, dimension(:,:,:), pointer mom_variables::accel_diag_ptrs::pfu => NULL() Zonal acceleration due to pressure forces [m s-2].
-
real, dimension(:,:,:), pointer mom_variables::accel_diag_ptrs::pfv => NULL() Meridional acceleration due to pressure forces [m s-2].
-
real, dimension(:,:,:), pointer mom_variables::accel_diag_ptrs::du_dt_visc => NULL() Zonal acceleration due to vertical viscosity [m s-2].
-
real, dimension(:,:,:), pointer mom_variables::accel_diag_ptrs::dv_dt_visc => NULL() Meridional acceleration due to vertical viscosity [m s-2].
-
real, dimension(:,:,:), pointer mom_variables::accel_diag_ptrs::du_dt_dia => NULL() Zonal acceleration due to diapycnal mixing [m s-2].
-
real, dimension(:,:,:), pointer mom_variables::accel_diag_ptrs::dv_dt_dia => NULL() Meridional acceleration due to diapycnal mixing [m s-2].
-
real, dimension(:,:,:), pointer mom_variables::accel_diag_ptrs::du_other => NULL() Zonal velocity changes due to any other processes that are not due to any explicit accelerations [m s-1].
-
real, dimension(:,:,:), pointer mom_variables::accel_diag_ptrs::dv_other => NULL() Meridional velocity changes due to any other processes that are not due to any explicit accelerations [m s-1].
-
real, dimension(:,:,:), pointer mom_variables::accel_diag_ptrs::gradkeu => NULL() gradKEu = - d/dx(u2) [m s-2]
-
real, dimension(:,:,:), pointer mom_variables::accel_diag_ptrs::gradkev => NULL() gradKEv = - d/dy(u2) [m s-2]
-
real, dimension(:,:,:), pointer mom_variables::accel_diag_ptrs::rv_x_v => NULL() rv_x_v = rv * v at u [m s-2]
-
real, dimension(:,:,:), pointer mom_variables::accel_diag_ptrs::rv_x_u => NULL() rv_x_u = rv * u at v [m s-2]
-
- type
Control structure for adaptive coordinates (coord_adapt).
Public Members
-
integer
nk¶ Number of layers/levels.
-
real, dimension(:), allocatable coord_adapt::adapt_cs::coordinateresolution Nominal near-surface resolution [H ~> m or kg m-2].
-
real
adapttimeratio¶ Ratio of optimisation and diffusion timescales.
-
real
adaptalpha¶ Nondimensional coefficient determining how much optimisation to apply.
-
real
adaptzoom¶ Near-surface zooming depth [H ~> m or kg m-2].
-
real
adaptzoomcoeff¶ Near-surface zooming coefficient.
-
real
adaptbuoycoeff¶ Stratification-dependent diffusion coefficient.
-
real
adaptdrho0¶ Reference density difference for stratification-dependent diffusion [kg m-3].
-
logical
adaptdomin= .false.¶ If true, form a HYCOM1-like mixed layet by preventing interfaces from becoming shallower than the depths set by coordinateResolution.
-
integer
- type
The control structure for the advect_test_tracer module.
Public Members
-
integer
ntr= NTR¶ Number of tracers in this module.
-
logical
coupled_tracers= .false.¶ These tracers are not offered to the coupler.
-
character(len=200) advection_test_tracer::advection_test_tracer_cs::tracer_ic_file The full path to the IC file, or ” ” to initialize internally.
-
type(time_type), pointer advection_test_tracer::advection_test_tracer_cs::time => NULL() A pointer to the ocean model’s clock.
-
type(tracer_registry_type), pointer advection_test_tracer::advection_test_tracer_cs::tr_reg => NULL() A pointer to the MOM tracer registry.
-
real, dimension(:,:,:,:), pointer advection_test_tracer::advection_test_tracer_cs::tr => NULL() The array of tracers used in this subroutine, in g m-3?
-
real, dimension(ntr) advection_test_tracer::advection_test_tracer_cs::land_val = -1.0 The value of tr used where land is masked out.
-
logical
use_sponge¶ If true, sponges may be applied somewhere in the domain.
-
logical
tracers_may_reinit¶ If true, the tracers may be set up via the initialization code if they are not found in the restart files. Otherwise it is a fatal error if the tracers are not found in the restart files of a restarted run.
-
real
x_origin¶ Parameters describing the test functions.
-
real
x_width¶ Parameters describing the test functions.
-
real
y_origin¶ Parameters describing the test functions.
-
real
y_width¶ Parameters describing the test functions.
-
integer, dimension(ntr) advection_test_tracer::advection_test_tracer_cs::ind_tr Indices returned by aof_set_coupler_flux if it is used and the surface tracer concentrations are to be provided to the coupler.
-
type(diag_ctrl), pointer advection_test_tracer::advection_test_tracer_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
type(mom_restart_cs), pointer advection_test_tracer::advection_test_tracer_cs::restart_csp => NULL() A pointer to the restart control structure.
-
type(vardesc), dimension(ntr) advection_test_tracer::advection_test_tracer_cs::tr_desc Descriptions and metadata for the tracers.
-
integer
- type
ALE control structure.
Public Members
-
logical
remap_uv_using_old_alg¶ If true, uses the old “remapping via a delta z” method. If False, uses the new method that remaps between grids described by h.
-
real
regrid_time_scale¶ The time-scale used in blending between the current (old) grid and the target (new) grid. (s)
-
type(regridding_cs) mom_ale::ale_cs::regridcs Regridding parameters and work arrays.
-
type(remapping_cs) mom_ale::ale_cs::remapcs Remapping parameters and work arrays.
-
integer
nk¶ Used only for queries, not directly by this module.
-
logical
remap_after_initialization¶ Indicates whether to regrid/remap after initializing the state.
-
logical
show_call_tree¶ For debugging.
-
type(diag_ctrl), pointer mom_ale::ale_cs::diag structure to regulate output
-
integer, dimension(:), allocatable mom_ale::ale_cs::id_tracer_remap_tendency diagnostic id
-
integer, dimension(:), allocatable mom_ale::ale_cs::id_htracer_remap_tendency diagnostic id
-
integer, dimension(:), allocatable mom_ale::ale_cs::id_htracer_remap_tendency_2d diagnostic id
-
logical, dimension(:), allocatable mom_ale::ale_cs::do_tendency_diag flag for doing diagnostics
-
integer
id_dzregrid= -1¶ diagnostic id
-
integer
id_u_preale= -1¶ diagnostic id for zonal velocity before ALE.
-
integer
id_v_preale= -1¶ diagnostic id for meridional velocity before ALE.
-
integer
id_h_preale= -1¶ diagnostic id for layer thicknesses before ALE.
-
integer
id_t_preale= -1¶ diagnostic id for temperatures before ALE.
-
integer
id_s_preale= -1¶ diagnostic id for salinities before ALE.
-
integer
id_e_preale= -1¶ diagnostic id for interface heights before ALE.
-
integer
id_vert_remap_h= -1¶ diagnostic id for layer thicknesses used for remapping
-
integer
id_vert_remap_h_tendency= -1¶ diagnostic id for layer thickness tendency due to ALE
-
logical
- type
ALE sponge control structure.
Public Members
-
integer
nz¶ The total number of layers.
-
integer
nz_data¶ The total number of arbritary layers (used by older code).
-
integer
isc¶ The starting i-index of the computational domain at h.
-
integer
iec¶ The ending i-index of the computational domain at h.
-
integer
jsc¶ The starting j-index of the computational domain at h.
-
integer
jec¶ The ending j-index of the computational domain at h.
-
integer
iscb¶ The starting I-index of the computational domain at u/v.
-
integer
iecb¶ The ending I-index of the computational domain at u/v.
-
integer
jscb¶ The starting J-index of the computational domain at u/v.
-
integer
jecb¶ The ending J-index of the computational domain at h.
-
integer
isd¶ The starting i-index of the data domain at h.
-
integer
ied¶ The ending i-index of the data domain at h.
-
integer
jsd¶ The starting j-index of the data domain at h.
-
integer
jed¶ The ending j-index of the data domain at h.
-
integer
num_col¶ The number of sponge tracer points within the computational domain.
-
integer
num_col_u¶ The number of sponge u-points within the computational domain.
-
integer
num_col_v¶ The number of sponge v-points within the computational domain.
-
integer
fldno= 0¶ The number of fields which have already been registered by calls to set_up_sponge_field.
-
logical
sponge_uv¶ Control whether u and v are included in sponge.
-
integer, dimension(:), pointer mom_ale_sponge::ale_sponge_cs::col_i => NULL() Array of the i-indicies of each tracer columns being damped.
-
integer, dimension(:), pointer mom_ale_sponge::ale_sponge_cs::col_j => NULL() Array of the j-indicies of each tracer columns being damped.
-
integer, dimension(:), pointer mom_ale_sponge::ale_sponge_cs::col_i_u => NULL() Array of the i-indicies of each u-columns being damped.
-
integer, dimension(:), pointer mom_ale_sponge::ale_sponge_cs::col_j_u => NULL() Array of the j-indicies of each u-columns being damped.
-
integer, dimension(:), pointer mom_ale_sponge::ale_sponge_cs::col_i_v => NULL() Array of the i-indicies of each v-columns being damped.
-
integer, dimension(:), pointer mom_ale_sponge::ale_sponge_cs::col_j_v => NULL() Array of the j-indicies of each v-columns being damped.
-
real, dimension(:), pointer mom_ale_sponge::ale_sponge_cs::iresttime_col => NULL() The inverse restoring time of each tracer column [s-1].
-
real, dimension(:), pointer mom_ale_sponge::ale_sponge_cs::iresttime_col_u => NULL() The inverse restoring time of each u-column [s-1].
-
real, dimension(:), pointer mom_ale_sponge::ale_sponge_cs::iresttime_col_v => NULL() The inverse restoring time of each v-column [s-1].
-
type(p3d), dimension( 50 ) mom_ale_sponge::ale_sponge_cs::var Pointers to the fields that are being damped.
-
type(p2d), dimension( 50 ) mom_ale_sponge::ale_sponge_cs::ref_val The values to which the fields are damped.
-
type(p2d) mom_ale_sponge::ale_sponge_cs::ref_val_u The values to which the u-velocities are damped.
-
type(p2d) mom_ale_sponge::ale_sponge_cs::ref_val_v The values to which the v-velocities are damped.
-
type(p3d) mom_ale_sponge::ale_sponge_cs::var_u Pointer to the u velocities. that are being damped.
-
type(p3d) mom_ale_sponge::ale_sponge_cs::var_v Pointer to the v velocities. that are being damped.
-
type(p2d) mom_ale_sponge::ale_sponge_cs::ref_h Grid on which reference data is provided (older code).
-
type(p2d) mom_ale_sponge::ale_sponge_cs::ref_hu u-point grid on which reference data is provided (older code).
-
type(p2d) mom_ale_sponge::ale_sponge_cs::ref_hv v-point grid on which reference data is provided (older code).
-
type(diag_ctrl), pointer mom_ale_sponge::ale_sponge_cs::diag A structure that is used to regulate the timing of diagnostic output.
-
type(remapping_cs) mom_ale_sponge::ale_sponge_cs::remap_cs Remapping parameters and work arrays.
-
logical
new_sponges¶ True if using newer sponge code.
-
integer
- interface
Copy the contents of one horizontal index type into another.
Private Functions
-
subroutine mom_hor_index::assignment(=)::hit_assign(HI1 HI1, HI2 HI2) HIT_assign copies one hor_index_type into another. It is accessed via an assignment (=) operator.
- Parameters
[out] hi1: Horizontal index type to copy to[in] hi2: Horizontal index type to copy from
-
- interface
Copy the value of one extended-fixed-point number into another.
Private Functions
-
subroutine mom_coms::assignment(=)::efp_assign(EFP1 EFP1, EFP2 EFP2) Copy one extended-fixed-point number into another.
- Parameters
[out] efp1: The recipient extended fixed point number[in] efp2: The source extended fixed point number
-
- type
A group of 1D axes that comprise a 1D/2D/3D mesh.
Public Members
-
character(len=15) mom_diag_mediator::axes_grp::id The id string for this particular combination of handles.
-
integer
rank¶ Number of dimensions in the list of axes.
-
integer, dimension(:), allocatable mom_diag_mediator::axes_grp::handles Handles to 1D axes.
-
type(diag_ctrl), pointer mom_diag_mediator::axes_grp::diag_cs => null() Circular link back to the main diagnostics control structure (Used to avoid passing said structure into every possible call).
-
character(len=9) mom_diag_mediator::axes_grp::x_cell_method = '' Default nature of data representation, if axes group includes x-direction.
-
character(len=9) mom_diag_mediator::axes_grp::y_cell_method = '' Default nature of data representation, if axes group includes y-direction.
-
character(len=9) mom_diag_mediator::axes_grp::v_cell_method = '' Default nature of data representation, if axes group includes vertical direction.
-
integer
nz= 0¶ Vertical dimension of diagnostic.
-
integer
vertical_coordinate_number= 0¶ Index of the corresponding diag_remap_ctrl for this axis group.
-
logical
is_h_point= .false.¶ If true, indicates that this axes group is for an h-point located field.
-
logical
is_q_point= .false.¶ If true, indicates that this axes group is for a q-point located field.
-
logical
is_u_point= .false.¶ If true, indicates that this axes group is for a u-point located field.
-
logical
is_v_point= .false.¶ If true, indicates that this axes group is for a v-point located field.
-
logical
is_layer= .false.¶ If true, indicates that this axes group is for a layer vertically-located field.
-
logical
is_interface= .false.¶ If true, indicates that this axes group is for an interface vertically-located field.
-
logical
is_native= .true.¶ If true, indicates that this axes group is for a native model grid. False for any other grid. Used for rank>2.
-
logical
needs_remapping= .false.¶ If true, indicates that this axes group is for a intensive layer-located field that must be remapped to these axes. Used for rank>2.
-
logical
needs_interpolating= .false.¶ If true, indicates that this axes group is for a sampled interface-located field that must be interpolated to these axes. Used for rank>2.
-
integer
downsample_level= 1¶ If greater than 1, the factor by which this diagnostic/axes/masks be downsampled.
-
type(axes_grp), pointer mom_diag_mediator::axes_grp::xyave_axes => null() The associated 1d axes for horizontall area-averaged diagnostics.
-
integer
id_area= -1¶ The diag_manager id for area to be used for cell_measure of variables with this axes_grp.
-
integer
id_volume= -1¶ The diag_manager id for volume to be used for cell_measure of variables with this axes_grp.
-
real, dimension(:,:), pointer mom_diag_mediator::axes_grp::mask2d => null() Mask for 2d (x-y) axes.
-
real, dimension(:,:,:), pointer mom_diag_mediator::axes_grp::mask3d => null() Mask for 3d axes.
-
type(diag_dsamp), dimension(2: 2 ) mom_diag_mediator::axes_grp::dsamp Downsample container.
-
- type
The barotropic stepping control stucture.
Unnamed Group
-
real, dimension( : , : , : ), allocatable mom_barotropic::barotropic_cs::frhatu The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
real, dimension( : , : , : ), allocatable mom_barotropic::barotropic_cs::frhatv The fraction of the total column thickness interpolated to v grid points in each layer [nondim].
-
real, dimension( : , : ), allocatable mom_barotropic::barotropic_cs::idatu Inverse of the basin depth at u grid points [Z-1 ~> m-1].
-
real, dimension( : , : ), allocatable mom_barotropic::barotropic_cs::lin_drag_u A spatially varying linear drag coefficient acting on the zonal barotropic flow [H T-1 ~> m s-1 or kg m-2 s-1].
-
real, dimension( : , : ), allocatable mom_barotropic::barotropic_cs::uhbt_ic The barotropic solvers estimate of the zonal transport as the initial condition for the next call to btstep [H L2 T-1 ~> m3 s-1 or kg s-1].
-
real, dimension( : , : ), allocatable mom_barotropic::barotropic_cs::ubt_ic The barotropic solvers estimate of the zonal velocity that will be the initial condition for the next call to btstep [L T-1 ~> m s-1].
-
real, dimension( : , : ), allocatable mom_barotropic::barotropic_cs::ubtav The barotropic zonal velocity averaged over the baroclinic time step [L T-1 ~> m s-1].
-
real, dimension( : , : ), allocatable mom_barotropic::barotropic_cs::idatv Inverse of the basin depth at v grid points [Z-1 ~> m-1].
-
real, dimension( : , : ), allocatable mom_barotropic::barotropic_cs::lin_drag_v A spatially varying linear drag coefficient acting on the zonal barotropic flow [H T-1 ~> m s-1 or kg m-2 s-1].
-
real, dimension( : , : ), allocatable mom_barotropic::barotropic_cs::vhbt_ic The barotropic solvers estimate of the zonal transport as the initial condition for the next call to btstep [H L2 T-1 ~> m3 s-1 or kg s-1].
-
real, dimension( : , : ), allocatable mom_barotropic::barotropic_cs::vbt_ic The barotropic solvers estimate of the zonal velocity that will be the initial condition for the next call to btstep [L T-1 ~> m s-1].
-
real, dimension( : , : ), allocatable mom_barotropic::barotropic_cs::vbtav The barotropic meridional velocity averaged over the baroclinic time step [L T-1 ~> m s-1].
-
real, dimension( : , : ), allocatable mom_barotropic::barotropic_cs::eta_cor The difference between the free surface height from the barotropic calculation and the sum of the layer thicknesses. This difference is imposed as a forcing term in the barotropic calculation over a baroclinic timestep [H ~> m or kg m-2].
-
real, dimension( : , : ), allocatable mom_barotropic::barotropic_cs::eta_cor_bound A limit on the rate at which eta_cor can be applied while avoiding instability [H T-1 ~> m s-1 or kg m-2 s-1]. This is only used if CSbound_BT_corr is true.
-
real, dimension( : , : ), allocatable mom_barotropic::barotropic_cs::ua_polarity Test vector components for checking grid polarity.
-
real, dimension( : , : ), allocatable mom_barotropic::barotropic_cs::va_polarity Test vector components for checking grid polarity.
-
real, dimension( : , : ), allocatable mom_barotropic::barotropic_cs::bathyt A copy of bathyT (ocean bottom depth) with wide halos [Z ~> m].
-
real, dimension( : , : ), allocatable mom_barotropic::barotropic_cs::iareat This is a copy of GIareaT with wide halos, but will still utilize the macro IareaT when referenced, [L-2 ~> m-2].
-
real, dimension( : , : ), allocatable mom_barotropic::barotropic_cs::d_u_cor A simply averaged depth at u points [Z ~> m].
-
real, dimension( : , : ), allocatable mom_barotropic::barotropic_cs::dy_cu A copy of Gdy_Cu with wide halos [L ~> m].
-
real, dimension( : , : ), allocatable mom_barotropic::barotropic_cs::idxcu A copy of GIdxCu with wide halos [L-1 ~> m-1].
-
real, dimension( : , : ), allocatable mom_barotropic::barotropic_cs::d_v_cor A simply averaged depth at v points [Z ~> m].
-
real, dimension( : , : ), allocatable mom_barotropic::barotropic_cs::dx_cv A copy of Gdx_Cv with wide halos [L ~> m].
-
real, dimension( : , : ), allocatable mom_barotropic::barotropic_cs::idycv A copy of GIdyCv with wide halos [L-1 ~> m-1].
-
real, dimension( : , : ), allocatable mom_barotropic::barotropic_cs::q_d f / D at PV points [Z-1 T-1 ~> m-1 s-1].
-
real, dimension(:,:,:), pointer mom_barotropic::barotropic_cs::frhatu1 => NULL() Predictor step values of frhatu stored for diagnostics.
-
real, dimension(:,:,:), pointer mom_barotropic::barotropic_cs::frhatv1 => NULL() Predictor step values of frhatv stored for diagnostics.
-
type(bt_obc_type) mom_barotropic::barotropic_cs::bt_obc A structure with all of this modules fields for applying open boundary conditions.
-
real
rho0¶ The density used in the Boussinesq approximation [kg m-3].
-
real
dtbt¶ The barotropic time step [s].
-
real
dtbt_fraction¶ The fraction of the maximum time-step that should used. The default is 0.98.
-
real
dtbt_max¶ The maximum stable barotropic time step [s].
-
real
dt_bt_filter¶ The time-scale over which the barotropic mode solutions are filtered [T ~> s] if positive, or as a fraction of DT if negative [nondim]. This can never be taken to be longer than 2*dt. Set this to 0 to apply no filtering.
-
integer
nstep_last= 0¶ The number of barotropic timesteps per baroclinic time step the last time btstep was called.
-
real
bebt¶ A nondimensional number, from 0 to 1, that determines the gravity wave time stepping scheme. 0.0 gives a forward-backward scheme, while 1.0 give backward Euler. In practice, bebt should be of order 0.2 or greater.
-
logical
split¶ If true, use the split time stepping scheme.
-
logical
bound_bt_corr¶ If true, the magnitude of the fake mass source in the barotropic equation that drives the two estimates of the free surface height toward each other is bounded to avoid driving corrective velocities that exceed MAXCFL_BT_CONT.
-
logical
gradual_bt_ics¶ If true, adjust the initial conditions for the barotropic solver to the values from the layered solution over a whole timestep instead of instantly. This is a decent approximation to the inclusion of sum(u dh_dt) while also correcting for truncation errors.
-
logical
sadourny¶ If true, the Coriolis terms are discretized with Sadourny’s energy conserving scheme, otherwise the Arakawa & Hsu scheme is used. If the deformation radius is not resolved Sadourny’s scheme should probably be used.
-
logical
nonlinear_continuity¶ If true, the barotropic continuity equation uses the full ocean thickness for transport.
-
integer
nonlin_cont_update_period¶ The number of barotropic time steps between updates to the face area, or 0 only to update at the start of a call to btstep. The default is 1.
-
logical
bt_project_velocity¶ If true, step the barotropic velocity first and project out the velocity tendency by 1+BEBT when calculating the transport. The default (false) is to use a predictor continuity step to find the pressure field, and then do a corrector continuity step using a weighted average of the old and new velocities, with weights of (1-BEBT) and BEBT.
-
logical
dynamic_psurf¶ If true, add a dynamic pressure due to a viscous ice shelf, for instance.
-
real
dmin_dyn_psurf¶ The minimum depth to use in limiting the size of the dynamic surface pressure for stability [Z ~> m].
-
real
ice_strength_length¶ The length scale at which the damping rate due to the ice strength should be the same as if a Laplacian were applied [L ~> m].
-
real
const_dyn_psurf¶ The constant that scales the dynamic surface pressure [nondim]. Stable values are < ~1.0. The default is 0.9.
-
logical
tides¶ If true, apply tidal momentum forcing.
-
real
g_extra¶ A nondimensional factor by which gtot is enhanced.
-
integer
hvel_scheme¶ An integer indicating how the thicknesses at velocity points are calculated. Valid values are given by the parameters defined below: HARMONIC, ARITHMETIC, HYBRID, and FROM_BT_CONT.
-
logical
strong_drag¶ If true, use a stronger estimate of the retarding effects of strong bottom drag.
-
logical
linear_wave_drag¶ If true, apply a linear drag to the barotropic velocities, using rates set by lin_drag_u & _v divided by the depth of the ocean.
-
logical
linearized_bt_pv¶ If true, the PV and interface thicknesses used in the barotropic Coriolis calculation is time invariant and linearized.
-
logical
use_wide_halos¶ If true, use wide halos and march in during the barotropic time stepping for efficiency.
-
logical
clip_velocity¶ If true, limit any velocity components that are are large enough for a CFL number to exceed CFL_trunc. This should only be used as a desperate debugging measure.
-
logical
debug¶ If true, write verbose checksums for debugging purposes.
-
logical
debug_bt¶ If true, write verbose checksums for debugging purposes.
-
real
vel_underflow¶ Velocity components smaller than vel_underflow are set to 0 [L T-1 ~> m s-1].
-
real
maxvel¶ Velocity components greater than maxvel are truncated to maxvel [m s-1].
-
real
cfl_trunc¶ If clip_velocity is true, velocity components will be truncated when they are large enough that the corresponding CFL number exceeds this value, nondim.
-
real
maxcfl_bt_cont¶ The maximum permitted CFL number associated with the barotropic accelerations from the summed velocities times the time-derivatives of thicknesses. The default is 0.1, and there will probably be real problems if this were set close to 1.
-
logical
bt_cont_bounds¶ If true, use the BT_cont_type variables to set limits on the magnitude of the corrective mass fluxes.
-
logical
visc_rem_u_uh0¶ If true, use the viscous remnants when estimating the barotropic velocities that were used to calculate uh0 and vh0. False is probably the better choice.
-
logical
adjust_bt_cont¶ If true, adjust the curve fit to the BT_cont type that is used by the barotropic solver to match the transport about which the flow is being linearized.
-
logical
use_old_coriolis_bracket_bug¶ If True, use an order of operations that is not bitwise rotationally symmetric in the meridional Coriolis term of the barotropic solver.
-
type(time_type), pointer mom_barotropic::barotropic_cs::time => NULL() A pointer to the ocean models clock.
-
type(diag_ctrl), pointer mom_barotropic::barotropic_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
type(mom_domain_type), pointer mom_barotropic::barotropic_cs::bt_domain => NULL() The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
type(hor_index_type), pointer mom_barotropic::barotropic_cs::debug_bt_hi => NULL() debugging copy of horizontal index_type
-
type(tidal_forcing_cs), pointer mom_barotropic::barotropic_cs::tides_csp => NULL() The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
logical
module_is_initialized= .false.¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
isdw¶ The lower i-memory limit for the wide halo arrays.
-
integer
iedw¶ The upper i-memory limit for the wide halo arrays.
-
integer
jsdw¶ The lower j-memory limit for the wide halo arrays.
-
integer
jedw¶ The upper j-memory limit for the wide halo arrays.
-
type(group_pass_type) mom_barotropic::barotropic_cs::pass_q_dcor Handle for a group halo pass.
-
type(group_pass_type) mom_barotropic::barotropic_cs::pass_gtot Handle for a group halo pass.
-
type(group_pass_type) mom_barotropic::barotropic_cs::pass_tmp_uv Handle for a group halo pass.
-
type(group_pass_type) mom_barotropic::barotropic_cs::pass_eta_bt_rem Handle for a group halo pass.
-
type(group_pass_type) mom_barotropic::barotropic_cs::pass_force_hbt0_cor_ref Handle for a group halo pass.
-
type(group_pass_type) mom_barotropic::barotropic_cs::pass_dat_uv Handle for a group halo pass.
-
type(group_pass_type) mom_barotropic::barotropic_cs::pass_eta_ubt Handle for a group halo pass.
-
type(group_pass_type) mom_barotropic::barotropic_cs::pass_etaav Handle for a group halo pass.
-
type(group_pass_type) mom_barotropic::barotropic_cs::pass_ubt_cor Handle for a group halo pass.
-
type(group_pass_type) mom_barotropic::barotropic_cs::pass_ubta_uhbta Handle for a group halo pass.
-
type(group_pass_type) mom_barotropic::barotropic_cs::pass_e_anom Handle for a group halo pass.
-
integer
id_pfu_bt= -1¶ Diagnostic IDs.
-
integer
id_pfv_bt= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_coru_bt= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_corv_bt= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_ubtforce= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_vbtforce= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_uaccel= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_vaccel= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_visc_rem_u= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_visc_rem_v= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_eta_cor= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_ubt= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_vbt= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_eta_bt= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_ubtav= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_vbtav= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_ubt_st= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_vbt_st= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_eta_st= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_ubt_hifreq= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_vbt_hifreq= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_eta_hifreq= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_uhbt_hifreq= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_vhbt_hifreq= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_eta_pred_hifreq= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_gtotn= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_gtots= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_gtote= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_gtotw= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_uhbt= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_frhatu= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_vhbt= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_frhatv= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_frhatu1= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_frhatv1= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_btc_fa_u_ee= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_btc_fa_u_e0= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_btc_fa_u_w0= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_btc_fa_u_ww= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_btc_ubt_ee= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_btc_ubt_ww= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_btc_fa_v_nn= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_btc_fa_v_n0= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_btc_fa_v_s0= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_btc_fa_v_ss= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_btc_vbt_nn= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_btc_vbt_ss= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_uhbt0= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
integer
id_vhbt0= -1¶ The fraction of the total column thickness interpolated to u grid points in each layer [nondim].
-
-
interface
bchksum¶ Checksums an array (2d or 3d) staggered at corner points.
Private Functions
-
subroutine
chksum_b_2d(array array, mesg mesg, HI HI, haloshift haloshift, symmetric symmetric, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums a 2d array staggered at corner points.
- Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] mesg: An identifying message[in] haloshift: The width of halos to check (default 0)[in] symmetric: If true, do the checksums on the full symmetric computational domain.[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
chksum_b_3d(array array, mesg mesg, HI HI, haloshift haloshift, symmetric symmetric, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums a 3d array staggered at corner points.
- Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] mesg: An identifying message[in] haloshift: The width of halos to check (default 0)[in] symmetric: If true, do the checksums on the full symmetric computational domain.[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
-
interface
bchksum_pair¶ Checksums a pair of arrays (2d or 3d) staggered at corner points.
Private Functions
-
subroutine
chksum_pair_b_2d(mesg mesg, arrayA arrayA, arrayB arrayB, HI HI, haloshift haloshift, symmetric symmetric, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums on a pair of 2d arrays staggered at q-points.
- Parameters
[in] mesg: Identifying messages[in] hi: A horizontal index type[in] arraya: The first array to be checksummed[in] arrayb: The second array to be checksummed[in] symmetric: If true, do the checksums on the full symmetric computational domain.[in] haloshift: The width of halos to check (default 0)[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
chksum_pair_b_3d(mesg mesg, arrayA arrayA, arrayB arrayB, HI HI, haloshift haloshift, symmetric symmetric, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums on a pair of 3d arrays staggered at q-points.
- Parameters
[in] mesg: Identifying messages[in] hi: A horizontal index type[in] arraya: The first array to be checksummed[in] arrayb: The second array to be checksummed[in] haloshift: The width of halos to check (default 0)[in] symmetric: If true, do the checksums on the full symmetric computational domain.[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
- type
Control structure for BFB_surface_forcing.
Public Members
-
logical
use_temperature¶ If true, temperature and salinity are used as state variables.
-
logical
restorebuoy¶ If true, use restoring surface buoyancy forcing.
-
real
rho0¶ The density used in the Boussinesq approximation [kg m-3].
-
real
g_earth¶ The gravitational acceleration [L2 Z-1 T-2 ~> m s-2].
-
real
flux_const¶ The restoring rate at the surface [m s-1].
-
real
gust_const¶ A constant unresolved background gustiness that contributes to ustar [Pa].
-
real
sst_s¶ SST at the southern edge of the linear forcing ramp [degC].
-
real
sst_n¶ SST at the northern edge of the linear forcing ramp [degC].
-
real
lfrslat¶ Southern latitude where the linear forcing ramp begins [degLat].
-
real
lfrnlat¶ Northern latitude where the linear forcing ramp ends [degLat].
-
real
drho_dt¶ Rate of change of density with temperature [kg m-3 degC-1]. Note that temperature is being used as a dummy variable here. All temperatures are converted into density.
-
type(diag_ctrl), pointer bfb_surface_forcing::bfb_surface_forcing_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
logical
- type
Control structure including parameters for this module.
Public Members
-
real
bryan_lewis_c1¶ The vertical diffusivity values for Bryan-Lewis profile at |z|=D [m2 s-1].
-
real
bryan_lewis_c2¶ The amplitude of variation in diffusivity for the Bryan-Lewis diffusivity profile [m2 s-1].
-
real
bryan_lewis_c3¶ The inverse length scale for transition region in the Bryan-Lewis diffusivity profile [m-1].
-
real
bryan_lewis_c4¶ The depth where diffusivity is Bryan_Lewis_bl1 in the Bryan-Lewis profile [m].
-
real
bckgrnd_vdc1¶ Background diffusivity (Ledwell) when horiz_varying_background=.true. [Z2 T-1 ~> m2 s-1].
-
real
bckgrnd_vdc_eq¶ Equatorial diffusivity (Gregg) when horiz_varying_background=.true. [Z2 T-1 ~> m2 s-1].
-
real
bckgrnd_vdc_psim¶ Max. PSI induced diffusivity (MacKinnon) when horiz_varying_background=.true. [Z2 T-1 ~> m2 s-1].
-
real
bckgrnd_vdc_banda¶ Banda Sea diffusivity (Gordon) when horiz_varying_background=.true. [Z2 T-1 ~> m2 s-1].
-
real
kd_min¶ minimum diapycnal diffusivity [Z2 T-1 ~> m2 s-1]
-
real
kd¶ interior diapycnal diffusivity [Z2 T-1 ~> m2 s-1]
-
real
omega¶ The Earth’s rotation rate [T-1 ~> s-1].
-
real
n0_2omega¶ ratio of the typical Buoyancy frequency to twice the Earth’s rotation period, used with the Henyey scaling from the mixing
-
real
prandtl_bkgnd¶ Turbulent Prandtl number used to convert vertical background diffusivity into viscosity.
-
real
kd_tanh_lat_scale¶ A nondimensional scaling for the range of diffusivities with Kd_tanh_lat_fn. Valid values are in the range of -2 to 2; 0.4 reproduces CM2M.
-
real
kdml¶ mixed layer diapycnal diffusivity [Z2 T-1 ~> m2 s-1] when bulkmixedlayer==.false.
-
real
hmix¶ mixed layer thickness [Z ~> m] when bulkmixedlayer==.false.
-
logical
kd_tanh_lat_fn¶ If true, use the tanh dependence of Kd_sfc on latitude, like GFDL CM2.1/CM2M. There is no physical justification for this form, and it can not be used with Henyey_IGW_background.
-
logical
bryan_lewis_diffusivity¶ If true, background vertical diffusivity uses Bryan-Lewis (1979) like tanh profile.
-
logical
horiz_varying_background¶ If true, apply vertically uniform, latitude-dependent background diffusivity, as described in Danabasoglu et al., 2012.
-
logical
henyey_igw_background¶ If true, use a simplified variant of the Henyey et al, JGR (1986) latitudinal scaling for the background diapycnal diffusivity, which gives a marked decrease in the diffusivity near the equator. The simplification here is to assume that the in-situ stratification is the same as the reference stratificaiton.
-
logical
henyey_igw_background_new¶ same as Henyey_IGW_background but incorporate the effect of stratification on TKE dissipation, e = f/f_0 * acosh(N/f) / acosh(N_0/f_0) * e_0 where e is the TKE dissipation, and N_0 and f_0 are the reference buoyancy frequency and inertial frequencies respectively. e_0 is the reference dissipation at (N_0,f_0). In the previous version, N=N_0. Additionally, the squared inverse relationship between diapycnal diffusivities and stratification is included:
kd = e/N^2
where kd is the diapycnal diffusivity. This approach assumes that work done against gravity is uniformly distributed throughout the column. Whereas, kd=kd_0*e, as in the original version, concentrates buoyancy work in regions of strong stratification.
-
logical
bulkmixedlayer¶ If true, a refined bulk mixed layer scheme is used.
-
logical
debug¶ If true, turn on debugging in this module.
-
type(diag_ctrl), pointer mom_bkgnd_mixing::bkgnd_mixing_cs::diag => NULL() A structure that regulates diagnostic output.
-
integer
id_kd_bkgnd= -1¶ Diagnotic IDs.
-
integer
id_kv_bkgnd= -1¶ Diagnostic IDs.
-
real, dimension(:,:), allocatable mom_bkgnd_mixing::bkgnd_mixing_cs::kd_sfc surface value of the diffusivity [Z2 T-1 ~> m2 s-1]
-
real, dimension(:,:,:), allocatable mom_bkgnd_mixing::bkgnd_mixing_cs::kd_bkgnd Background diffusivity [Z2 T-1 ~> m2 s-1].
-
real, dimension(:,:,:), allocatable mom_bkgnd_mixing::bkgnd_mixing_cs::kv_bkgnd Background viscosity [Z2 s-1 ~> m2 s-1].
-
character(len=40) mom_bkgnd_mixing::bkgnd_mixing_cs::bkgnd_scheme_str = "none" Background scheme identifier.
-
real
- type
The control structure for the boundary impulse tracer package.
Public Members
-
integer
ntr= NTR_MAX¶ The number of tracers that are actually used.
-
logical
coupled_tracers= .false.¶ These tracers are not offered to the coupler.
-
type(time_type), pointer boundary_impulse_tracer::boundary_impulse_tracer_cs::time => NULL() A pointer to the ocean model’s clock.
-
type(tracer_registry_type), pointer boundary_impulse_tracer::boundary_impulse_tracer_cs::tr_reg => NULL() A pointer to the tracer registry.
-
real, dimension(:,:,:,:), pointer boundary_impulse_tracer::boundary_impulse_tracer_cs::tr => NULL() The array of tracers used in this subroutine, in g m-3?
-
logical
tracers_may_reinit¶ If true, boundary_impulse can be initialized if not found in restart file.
-
integer, dimension(ntr_max) boundary_impulse_tracer::boundary_impulse_tracer_cs::ind_tr Indices returned by aof_set_coupler_flux if it is used and the surface tracer concentrations are to be provided to the coupler.
-
integer
nkml¶ Number of layers in mixed layer.
-
real, dimension(ntr_max) boundary_impulse_tracer::boundary_impulse_tracer_cs::land_val = -1.0 A value to use to fill in tracers over land.
-
real
kw_eff¶ An effective piston velocity used to flux tracer out at the surface.
-
real
remaining_source_time¶ How much longer (same units as the timestep) to inject the tracer at the surface.
-
type(diag_ctrl), pointer boundary_impulse_tracer::boundary_impulse_tracer_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
type(mom_restart_cs), pointer boundary_impulse_tracer::boundary_impulse_tracer_cs::restart_csp => NULL() A pointer to the retart control structure.
-
type(vardesc), dimension(ntr_max) boundary_impulse_tracer::boundary_impulse_tracer_cs::tr_desc Descriptions and metadata for the tracers.
-
integer
- type
Container for information about the summed layer transports and how they will vary as the barotropic velocity is changed.
Public Members
-
real, dimension(:,:), allocatable mom_variables::bt_cont_type::fa_u_ee The effective open face area for zonal barotropic transport drawing from locations far to the east [H L ~> m2 or kg m-1].
-
real, dimension(:,:), allocatable mom_variables::bt_cont_type::fa_u_e0 The effective open face area for zonal barotropic transport drawing from nearby to the east [H L ~> m2 or kg m-1].
-
real, dimension(:,:), allocatable mom_variables::bt_cont_type::fa_u_w0 The effective open face area for zonal barotropic transport drawing from nearby to the west [H L ~> m2 or kg m-1].
-
real, dimension(:,:), allocatable mom_variables::bt_cont_type::fa_u_ww The effective open face area for zonal barotropic transport drawing from locations far to the west [H L ~> m2 or kg m-1].
-
real, dimension(:,:), allocatable mom_variables::bt_cont_type::ubt_ww uBT_WW is the barotropic velocity [L T-1 ~> m s-1], beyond which the marginal open face area is FA_u_WW. uBT_WW must be non-negative.
-
real, dimension(:,:), allocatable mom_variables::bt_cont_type::ubt_ee uBT_EE is a barotropic velocity [L T-1 ~> m s-1], beyond which the marginal open face area is FA_u_EE. uBT_EE must be non-positive.
-
real, dimension(:,:), allocatable mom_variables::bt_cont_type::fa_v_nn The effective open face area for meridional barotropic transport drawing from locations far to the north [H L ~> m2 or kg m-1].
-
real, dimension(:,:), allocatable mom_variables::bt_cont_type::fa_v_n0 The effective open face area for meridional barotropic transport drawing from nearby to the north [H L ~> m2 or kg m-1].
-
real, dimension(:,:), allocatable mom_variables::bt_cont_type::fa_v_s0 The effective open face area for meridional barotropic transport drawing from nearby to the south [H L ~> m2 or kg m-1].
-
real, dimension(:,:), allocatable mom_variables::bt_cont_type::fa_v_ss The effective open face area for meridional barotropic transport drawing from locations far to the south [H L ~> m2 or kg m-1].
-
real, dimension(:,:), allocatable mom_variables::bt_cont_type::vbt_ss vBT_SS is the barotropic velocity, [L T-1 ~> m s-1], beyond which the marginal open face area is FA_v_SS. vBT_SS must be non-negative.
-
real, dimension(:,:), allocatable mom_variables::bt_cont_type::vbt_nn vBT_NN is the barotropic velocity, [L T-1 ~> m s-1], beyond which the marginal open face area is FA_v_NN. vBT_NN must be non-positive.
-
real, dimension(:,:,:), allocatable mom_variables::bt_cont_type::h_u An effective thickness at zonal faces [H ~> m or kg m-2].
-
real, dimension(:,:,:), allocatable mom_variables::bt_cont_type::h_v An effective thickness at meridional faces [H ~> m or kg m-2].
-
type(group_pass_type) mom_variables::bt_cont_type::pass_polarity_bt Structure for polarity group halo updates.
-
type(group_pass_type) mom_variables::bt_cont_type::pass_fa_uv Structure for face area group halo updates.
-
- type
The barotropic stepping open boundary condition type.
Unnamed Group
-
integer
is_u_obc¶ Index ranges for the open boundary conditions.
-
integer
ie_u_obc¶ Index ranges for the open boundary conditions.
-
integer
js_u_obc¶ Index ranges for the open boundary conditions.
-
integer
je_u_obc¶ Index ranges for the open boundary conditions.
-
integer
is_v_obc¶ Index ranges for the open boundary conditions.
-
integer
ie_v_obc¶ Index ranges for the open boundary conditions.
-
integer
js_v_obc¶ Index ranges for the open boundary conditions.
-
integer
je_v_obc¶ Index ranges for the open boundary conditions.
-
logical
is_alloced= .false.¶ True if BT_OBC is in use and has been allocated.
-
type(group_pass_type) mom_barotropic::bt_obc_type::pass_uv Structure for group halo pass.
-
type(group_pass_type) mom_barotropic::bt_obc_type::pass_uhvh Structure for group halo pass.
-
type(group_pass_type) mom_barotropic::bt_obc_type::pass_h Structure for group halo pass.
-
type(group_pass_type) mom_barotropic::bt_obc_type::pass_cg Structure for group halo pass.
-
type(group_pass_type) mom_barotropic::bt_obc_type::pass_eta_outer Structure for group halo pass.
Private Members
-
real, dimension(:,:), pointer mom_barotropic::bt_obc_type::cg_u => NULL() The external wave speed at u-points [L T-1 ~> m s-1].
-
real, dimension(:,:), pointer mom_barotropic::bt_obc_type::cg_v => NULL() The external wave speed at u-points [L T-1 ~> m s-1].
-
real, dimension(:,:), pointer mom_barotropic::bt_obc_type::h_u => NULL() The total thickness at the u-points [H ~> m or kg m-2].
-
real, dimension(:,:), pointer mom_barotropic::bt_obc_type::h_v => NULL() The total thickness at the v-points [H ~> m or kg m-2].
-
real, dimension(:,:), pointer mom_barotropic::bt_obc_type::uhbt => NULL() The zonal barotropic thickness fluxes specified for open boundary conditions (if any) [H L2 T-1 ~> m3 s-1 or kg s-1].
-
real, dimension(:,:), pointer mom_barotropic::bt_obc_type::vhbt => NULL() The meridional barotropic thickness fluxes specified for open boundary conditions (if any) [H L2 T-1 ~> m3 s-1 or kg s-1].
-
real, dimension(:,:), pointer mom_barotropic::bt_obc_type::ubt_outer => NULL() The zonal velocities just outside the domain, as set by the open boundary conditions [L T-1 ~> m s-1].
-
real, dimension(:,:), pointer mom_barotropic::bt_obc_type::vbt_outer => NULL() The meridional velocities just outside the domain, as set by the open boundary conditions [L T-1 ~> m s-1].
-
real, dimension(:,:), pointer mom_barotropic::bt_obc_type::eta_outer_u => NULL() The surface height outside of the domain at a u-point with an open boundary condition [H ~> m or kg m-2].
-
real, dimension(:,:), pointer mom_barotropic::bt_obc_type::eta_outer_v => NULL() The surface height outside of the domain at a v-point with an open boundary condition [H ~> m or kg m-2].
-
logical
apply_u_obcs¶ True if this PE has an open boundary at a u-point.
-
logical
apply_v_obcs¶ True if this PE has an open boundary at a v-point.
-
integer
- type
The control structure with parameters for the MOM_bulk_mixed_layer module.
Unnamed Group
-
integer
id_ml_depth= -1¶ Diagnostic IDs.
-
integer
id_tke_wind= -1¶ Diagnostic IDs.
-
integer
id_tke_mixing= -1¶ Diagnostic IDs.
-
integer
id_tke_ribulk= -1¶ Diagnostic IDs.
-
integer
id_tke_conv= -1¶ Diagnostic IDs.
-
integer
id_tke_pen_sw= -1¶ Diagnostic IDs.
-
integer
id_tke_mech_decay= -1¶ Diagnostic IDs.
-
integer
id_tke_conv_decay= -1¶ Diagnostic IDs.
-
integer
id_tke_conv_s2= -1¶ Diagnostic IDs.
-
integer
id_pe_detrain= -1¶ Diagnostic IDs.
-
integer
id_pe_detrain2= -1¶ Diagnostic IDs.
-
integer
id_h_mismatch= -1¶ Diagnostic IDs.
-
integer
id_hsfc_used= -1¶ Diagnostic IDs.
-
integer
id_hsfc_max= -1¶ Diagnostic IDs.
-
integer
id_hsfc_min= -1¶ Diagnostic IDs.
Public Members
-
integer
nkml¶ The number of layers in the mixed layer.
-
integer
nkbl¶ The number of buffer layers.
-
integer
nsw¶ The number of bands of penetrating shortwave radiation.
-
real
mstar¶ The ratio of the friction velocity cubed to the TKE input to the mixed layer, nondimensional.
-
real
nstar¶ The fraction of the TKE input to the mixed layer available to drive entrainment [nondim].
-
real
nstar2¶ The fraction of potential energy released by convective adjustment that drives entrainment [nondim].
-
logical
absorb_all_sw¶ If true, all shortwave radiation is absorbed by the ocean, instead of passing through to the bottom mud.
-
real
tke_decay¶ The ratio of the natural Ekman depth to the TKE decay scale, nondimensional.
-
real
bulk_ri_ml¶ The efficiency with which mean kinetic energy released by mechanically forced entrainment of the mixed layer is converted to TKE [nondim].
-
real
bulk_ri_convective¶ The efficiency with which convectively released mean kinetic energy becomes TKE [nondim].
-
real
hmix_min¶ The minimum mixed layer thickness [H ~> m or kg m-2].
-
real
h_limit_fluxes¶ When the total ocean depth is less than this value [H ~> m or kg m-2], scale away all surface forcing to avoid boiling the ocean.
-
real
ustar_min¶ A minimum value of ustar to avoid numerical problems [Z T-1 ~> m s-1]. If the value is small enough, this should not affect the solution.
-
real
omega¶ The Earth’s rotation rate [T-1 ~> s-1].
-
real
dt_ds_wt¶ When forced to extrapolate T & S to match the layer densities, this factor (in degC / ppt) is combined with the derivatives of density with T & S to determines what direction is orthogonal to density contours. It should be a typical value of (dR/dS) / (dR/dT) in oceanic profiles. 6 degC ppt-1 might be reasonable.
-
real
hbuffer_min¶ The minimum buffer layer thickness when the mixed layer is very large [H ~> m or kg m-2].
-
real
hbuffer_rel_min¶ The minimum buffer layer thickness relative to the combined mixed and buffer layer thicknesses when they are thin [nondim].
-
real
bl_detrain_time¶ A timescale that characterizes buffer layer detrainment events [T ~> s].
-
real
bl_extrap_lim¶ A limit on the density range over which extrapolation can occur when detraining from the buffer layers, relative to the density range within the mixed and buffer layers, when the detrainment is going into the lightest interior layer [nondim].
-
real
bl_split_rho_tol¶ The fractional tolerance for matching layer target densities when splitting layers to deal with massive interior layers that are lighter than one of the mixed or buffer layers [nondim].
-
logical
ml_resort¶ If true, resort the layers by density, rather than doing convective adjustment.
-
integer
ml_presort_nz_conv_adj¶ If ML_resort is true, do convective adjustment on this many layers (starting from the top) before sorting the remaining layers.
-
real
omega_frac¶ When setting the decay scale for turbulence, use this fraction of the absolute rotation rate blended with the local value of f, as sqrt((1-of)*f^2 + of*4*omega^2).
-
logical
correct_absorption¶ If true, the depth at which penetrating shortwave radiation is absorbed is corrected by moving some of the heating upward in the water column. The default is false.
-
logical
resolve_ekman¶ If true, the nkml layers in the mixed layer are chosen to optimally represent the impact of the Ekman transport on the mixed layer TKE budget.
-
type(time_type), pointer mom_bulk_mixed_layer::bulkmixedlayer_cs::time => NULL() A pointer to the ocean model’s clock.
-
logical
tke_diagnostics= .false.¶ If true, calculate extensive diagnostics of the TKE budget.
-
logical
do_rivermix= .false.¶ Provide additional TKE to mix river runoff at the river mouths to rivermix_depth.
-
real
rivermix_depth= 0.0¶ The depth of mixing if do_rivermix is true [Z ~> m].
-
logical
limit_det¶ If true, limit the extent of buffer layer detrainment to be consistent with neighbors.
-
real
lim_det_dh_sfc¶ The fractional limit in the change between grid points of the surface region (mixed & buffer layer) thickness [nondim]. 0.5 by default.
-
real
lim_det_dh_bathy¶ The fraction of the total depth by which the thickness of the surface region (mixed & buffer layer) is allowed to change between grid points. Nondimensional, 0.2 by default.
-
logical
use_river_heat_content¶ If true, use the fluxesrunoff_Hflx field to set the heat carried by runoff, instead of using SST for temperature of liq_runoff.
-
logical
use_calving_heat_content¶ Use SST for temperature of froz_runoff.
-
logical
salt_reject_below_ml¶ It true, add salt below mixed layer (layer mode only)
-
type(diag_ctrl), pointer mom_bulk_mixed_layer::bulkmixedlayer_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
real
allowed_t_chg¶ The amount by which temperature is allowed to exceed previous values during detrainment, K.
-
real
allowed_s_chg¶ The amount by which salinity is allowed to exceed previous values during detrainment, ppt.
-
real, dimension(:,:), allocatable mom_bulk_mixed_layer::bulkmixedlayer_cs::ml_depth The mixed layer depth [H ~> m or kg m-2].
-
real, dimension(:,:), allocatable mom_bulk_mixed_layer::bulkmixedlayer_cs::diag_tke_wind The wind source of TKE.
-
real, dimension(:,:), allocatable mom_bulk_mixed_layer::bulkmixedlayer_cs::diag_tke_ribulk The resolved KE source of TKE.
-
real, dimension(:,:), allocatable mom_bulk_mixed_layer::bulkmixedlayer_cs::diag_tke_conv The convective source of TKE.
-
real, dimension(:,:), allocatable mom_bulk_mixed_layer::bulkmixedlayer_cs::diag_tke_pen_sw The TKE sink required to mix penetrating shortwave heating.
-
real, dimension(:,:), allocatable mom_bulk_mixed_layer::bulkmixedlayer_cs::diag_tke_mech_decay The decay of mechanical TKE.
-
real, dimension(:,:), allocatable mom_bulk_mixed_layer::bulkmixedlayer_cs::diag_tke_conv_decay The decay of convective TKE.
-
real, dimension(:,:), allocatable mom_bulk_mixed_layer::bulkmixedlayer_cs::diag_tke_mixing The work done by TKE to deepen the mixed layer.
-
real, dimension(:,:), allocatable mom_bulk_mixed_layer::bulkmixedlayer_cs::diag_tke_conv_s2 The convective source of TKE due to to mixing in sigma2.
-
real, dimension(:,:), allocatable mom_bulk_mixed_layer::bulkmixedlayer_cs::diag_pe_detrain The spurious source of potential energy due to mixed layer.
-
real, dimension(:,:), allocatable mom_bulk_mixed_layer::bulkmixedlayer_cs::diag_pe_detrain2 The spurious source of potential energy due to mixed layer only.
-
logical
allow_clocks_in_omp_loops¶ If true, clocks can be called from inside loops that can be threaded. To run with multiple threads, set to False.
-
type(group_pass_type) mom_bulk_mixed_layer::bulkmixedlayer_cs::pass_h_sum_hmbl_prev For group halo pass.
-
integer
-
interface
calculate_density¶ Calculates density of sea water from T, S and P.
Private Functions
-
subroutine
calculate_density_scalar(T T, S S, pressure pressure, rho rho, EOS EOS, rho_ref rho_ref)¶ Calls the appropriate subroutine to calculate density of sea water for scalar inputs. If rho_ref is present, the anomaly with respect to rho_ref is returned.
- Parameters
[in] t: Potential temperature referenced to the surface [degC][in] s: Salinity [ppt][in] pressure: Pressure [Pa][out] rho: Density (in-situ if pressure is local) [kg m-3]eos: Equation of state structure[in] rho_ref: A reference density [kg m-3].
-
subroutine
calculate_density_array(T T, S S, pressure pressure, rho rho, start start, npts npts, EOS EOS, rho_ref rho_ref)¶ Calls the appropriate subroutine to calculate the density of sea water for 1-D array inputs. If rho_ref is present, the anomaly with respect to rho_ref is returned.
- Parameters
[in] t: Potential temperature referenced to the surface [degC][in] s: Salinity [ppt][in] pressure: Pressure [Pa][out] rho: Density (in-situ if pressure is local) [kg m-3][in] start: Start index for computation[in] npts: Number of point to computeeos: Equation of state structure[in] rho_ref: A reference density [kg m-3].
-
subroutine
-
interface
calculate_density_derivs¶ Calculate the derivatives of density with temperature and salinity from T, S, and P.
Private Functions
-
subroutine
calculate_density_derivs_scalar(T T, S S, pressure pressure, drho_dT drho_dT, drho_dS drho_dS, EOS EOS)¶ Calls the appropriate subroutines to calculate density derivatives by promoting a scalar to a one-element array.
- Parameters
[in] t: Potential temperature referenced to the surface [degC][in] s: Salinity [ppt][in] pressure: Pressure [Pa][out] drho_dt: The partial derivative of density with potential temperature [kg m-3 degC-1].[out] drho_ds: The partial derivative of density with salinity, in [kg m-3 ppt-1].eos: Equation of state structure
-
subroutine
calculate_density_derivs_array(T T, S S, pressure pressure, drho_dT drho_dT, drho_dS drho_dS, start start, npts npts, EOS EOS)¶ Calls the appropriate subroutine to calculate density derivatives for 1-D array inputs.
- Parameters
[in] t: Potential temperature referenced to the surface [degC][in] s: Salinity [ppt][in] pressure: Pressure [Pa][out] drho_dt: The partial derivative of density with potential temperature [kg m-3 degC-1].[out] drho_ds: The partial derivative of density with salinity, in [kg m-3 ppt-1].[in] start: Starting index within the array[in] npts: The number of values to calculateeos: Equation of state structure
-
subroutine
-
interface
calculate_density_derivs_linear¶ For a given thermodynamic state, return the derivatives of density with temperature and salinity using the simple linear equation of state.
Private Functions
-
subroutine
calculate_density_derivs_scalar_linear(T T, S S, pressure pressure, drho_dT_out drho_dT_out, drho_dS_out drho_dS_out, Rho_T0_S0 Rho_T0_S0, dRho_dT dRho_dT, dRho_dS dRho_dS)¶ This subroutine calculates the partial derivatives of density * with potential temperature and salinity for a single point.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: pressure [Pa].[out] drho_dt_out: The partial derivative of density with potential temperature [kg m-3 degC-1].[out] drho_ds_out: The partial derivative of density with salinity [kg m-3 ppt-1].[in] rho_t0_s0: The density at T=0, S=0 [kg m-3].[in] drho_dt: The derivatives of density with temperature [kg m-3 degC-1].[in] drho_ds: The derivatives of density with salinity [kg m-3 ppt-1].
-
subroutine
calculate_density_derivs_array_linear(T T, S S, pressure pressure, drho_dT_out drho_dT_out, drho_dS_out drho_dS_out, Rho_T0_S0 Rho_T0_S0, dRho_dT dRho_dT, dRho_dS dRho_dS, start start, npts npts)¶ This subroutine calculates the partial derivatives of density * with potential temperature and salinity.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: Pressure [Pa].[out] drho_dt_out: The partial derivative of density with potential temperature [kg m-3 degC-1].[out] drho_ds_out: The partial derivative of density with salinity [kg m-3 ppt-1].[in] rho_t0_s0: The density at T=0, S=0 [kg m-3].[in] drho_dt: The derivative of density with temperature [kg m-3 degC-1].[in] drho_ds: The derivative of density with salinity [kg m-3 ppt-1].[in] start: The starting point in the arrays.[in] npts: The number of values to calculate.
-
subroutine
-
interface
calculate_density_derivs_nemo¶ For a given thermodynamic state, return the derivatives of density with conservative temperature and absolute salinity, the expressions derived for use with NEMO.
Private Functions
-
subroutine
calculate_density_derivs_scalar_nemo(T T, S S, pressure pressure, drho_dt drho_dt, drho_ds drho_ds)¶ Wrapper to calculate_density_derivs_array for scalar inputs.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [g kg-1].[in] pressure: Pressure [Pa].[out] drho_dt: The partial derivative of density with potential temperature [kg m-3 degC-1].[out] drho_ds: The partial derivative of density with salinity, in [kg m-3 ppt-1].
-
subroutine
calculate_density_derivs_array_nemo(T T, S S, pressure pressure, drho_dT drho_dT, drho_dS drho_dS, start start, npts npts)¶ For a given thermodynamic state, calculate the derivatives of density with conservative temperature and absolute salinity, using the expressions derived for use with NEMO.
- Parameters
[in] t: Conservative temperature [degC].[in] s: Absolute salinity [g kg-1].[in] pressure: pressure [Pa].[out] drho_dt: The partial derivative of density with potential temperature [kg m-3 degC-1].[out] drho_ds: The partial derivative of density with salinity, in [kg m-3 ppt-1].[in] start: The starting point in the arrays.[in] npts: The number of values to calculate.
-
subroutine
-
interface
calculate_density_derivs_teos10¶ For a given thermodynamic state, return the derivatives of density with conservative temperature and absolute salinity, using the TEOS10 expressions.
Private Functions
-
subroutine
calculate_density_derivs_scalar_teos10(T T, S S, pressure pressure, drho_dT drho_dT, drho_dS drho_dS)¶ For a given thermodynamic state, calculate the derivatives of density with conservative temperature and absolute salinity, using the TEOS10 expressions.
- Parameters
[in] t: Conservative temperature [degC][in] s: Absolute Salinity [g kg-1][in] pressure: pressure [Pa].[out] drho_dt: The partial derivative of density with conservative temperature [kg m-3 degC-1].[out] drho_ds: The partial derivative of density with absolute salinity, [kg m-3 (g/kg)-1].
-
subroutine
calculate_density_derivs_array_teos10(T T, S S, pressure pressure, drho_dT drho_dT, drho_dS drho_dS, start start, npts npts)¶ For a given thermodynamic state, calculate the derivatives of density with conservative temperature and absolute salinity, using the TEOS10 expressions.
- Parameters
[in] t: Conservative temperature [degC].[in] s: Absolute salinity [g kg-1].[in] pressure: pressure [Pa].[out] drho_dt: The partial derivative of density with conservative temperature [kg m-3 degC-1].[out] drho_ds: The partial derivative of density with absolute salinity, [kg m-3 (g/kg)-1].[in] start: The starting point in the arrays.[in] npts: The number of values to calculate.
-
subroutine
-
interface
calculate_density_derivs_wright¶ For a given thermodynamic state, return the derivatives of density with temperature and salinity.
Private Functions
-
subroutine
calculate_density_derivs_scalar_wright(T T, S S, pressure pressure, drho_dT drho_dT, drho_dS drho_dS)¶ The scalar version of calculate_density_derivs which promotes scalar inputs to a 1-element array and then demotes the output back to a scalar.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: pressure [Pa].[out] drho_dt: The partial derivative of density with potential temperature [kg m-3 degC-1].[out] drho_ds: The partial derivative of density with salinity, in [kg m-3 PSU-1].
-
subroutine
calculate_density_derivs_array_wright(T T, S S, pressure pressure, drho_dT drho_dT, drho_dS drho_dS, start start, npts npts)¶ For a given thermodynamic state, return the thermal/haline expansion coefficients.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: pressure [Pa].[out] drho_dt: The partial derivative of density with potential temperature [kg m-3 degC-1].[out] drho_ds: The partial derivative of density with salinity, in [kg m-3 PSU-1].[in] start: The starting point in the arrays.[in] npts: The number of values to calculate.
-
subroutine
-
interface
calculate_density_linear¶ Compute the density of sea water (in kg/m^3), or its anomaly from a reference density, using a simple linear equation of state from salinity (in psu), potential temperature (in deg C) and pressure [Pa].
Private Functions
-
subroutine
calculate_density_scalar_linear(T T, S S, pressure pressure, rho rho, Rho_T0_S0 Rho_T0_S0, dRho_dT dRho_dT, dRho_dS dRho_dS, rho_ref rho_ref)¶ This subroutine computes the density of sea water with a trivial linear equation of state (in [kg m-3]) from salinity (sal [PSU]), potential temperature (T [degC]), and pressure [Pa].
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: pressure [Pa].[out] rho: In situ density [kg m-3].[in] rho_t0_s0: The density at T=0, S=0 [kg m-3].[in] drho_dt: The derivatives of density with temperature [kg m-3 degC-1].[in] drho_ds: The derivatives of density with salinity in [kg m-3 ppt-1].[in] rho_ref: A reference density [kg m-3].
-
subroutine
calculate_density_array_linear(T T, S S, pressure pressure, rho rho, start start, npts npts, Rho_T0_S0 Rho_T0_S0, dRho_dT dRho_dT, dRho_dS dRho_dS, rho_ref rho_ref)¶ This subroutine computes the density of sea water with a trivial linear equation of state (in kg/m^3) from salinity (sal in psu), potential temperature (T [degC]), and pressure [Pa].
- Parameters
[in] t: potential temperature relative to the surface [degC].[in] s: salinity [PSU].[in] pressure: pressure [Pa].[out] rho: in situ density [kg m-3].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.[in] rho_t0_s0: The density at T=0, S=0 [kg m-3].[in] drho_dt: The derivatives of density with temperature [kg m-3 degC-1].[in] drho_ds: The derivatives of density with salinity in [kg m-3 ppt-1].[in] rho_ref: A reference density [kg m-3].
-
subroutine
-
interface
calculate_density_nemo¶ Compute the in situ density of sea water ([kg m-3]), or its anomaly with respect to a reference density, from absolute salinity (g/kg), conservative temperature (in deg C), and pressure [Pa], using the expressions derived for use with NEMO.
Private Functions
-
subroutine
calculate_density_scalar_nemo(T T, S S, pressure pressure, rho rho, rho_ref rho_ref)¶ This subroutine computes the in situ density of sea water (rho in [kg m-3]) from absolute salinity (S [g kg-1]), conservative temperature (T [degC]), and pressure [Pa]. It uses the expressions derived for use with NEMO.
- Parameters
[in] t: Conservative temperature [degC].[in] s: Absolute salinity [g kg-1].[in] pressure: pressure [Pa].[out] rho: In situ density [kg m-3].[in] rho_ref: A reference density [kg m-3].
-
subroutine
calculate_density_array_nemo(T T, S S, pressure pressure, rho rho, start start, npts npts, rho_ref rho_ref)¶ This subroutine computes the in situ density of sea water (rho in [kg m-3]) from absolute salinity (S [g kg-1]), conservative temperature (T [degC]), and pressure [Pa]. It uses the expressions derived for use with NEMO.
- Parameters
[in] t: Conservative temperature [degC].[in] s: Absolute salinity [g kg-1].[in] pressure: pressure [Pa].[out] rho: in situ density [kg m-3].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.[in] rho_ref: A reference density [kg m-3].
-
subroutine
-
interface
calculate_density_second_derivs¶ Calculates the second derivatives of density with various combinations of temperature, salinity, and pressure from T, S and P.
Private Functions
-
subroutine
calculate_density_second_derivs_scalar(T T, S S, pressure pressure, drho_dS_dS drho_dS_dS, drho_dS_dT drho_dS_dT, drho_dT_dT drho_dT_dT, drho_dS_dP drho_dS_dP, drho_dT_dP drho_dT_dP, EOS EOS)¶ Calls the appropriate subroutine to calculate density second derivatives for scalar nputs.
- Parameters
[in] t: Potential temperature referenced to the surface [degC][in] s: Salinity [ppt][in] pressure: Pressure [Pa][out] drho_ds_ds: Partial derivative of beta with respect to S [kg m-3 ppt-2][out] drho_ds_dt: Partial derivative of beta with respcct to T [kg m-3 ppt-1 degC-1][out] drho_dt_dt: Partial derivative of alpha with respect to T [kg m-3 degC-2][out] drho_ds_dp: Partial derivative of beta with respect to pressure [kg m-3 ppt-1 Pa-1][out] drho_dt_dp: Partial derivative of alpha with respect to pressure [kg m-3 degC-1 Pa-1]eos: Equation of state structure
-
subroutine
calculate_density_second_derivs_array(T T, S S, pressure pressure, drho_dS_dS drho_dS_dS, drho_dS_dT drho_dS_dT, drho_dT_dT drho_dT_dT, drho_dS_dP drho_dS_dP, drho_dT_dP drho_dT_dP, start start, npts npts, EOS EOS)¶ Calls the appropriate subroutine to calculate density second derivatives for 1-D array inputs.
- Parameters
[in] t: Potential temperature referenced to the surface [degC][in] s: Salinity [ppt][in] pressure: Pressure [Pa][out] drho_ds_ds: Partial derivative of beta with respect to S [kg m-3 ppt-2][out] drho_ds_dt: Partial derivative of beta with respcct to T [kg m-3 ppt-1 degC-1][out] drho_dt_dt: Partial derivative of alpha with respect to T [kg m-3 degC-2][out] drho_ds_dp: Partial derivative of beta with respect to pressure [kg m-3 ppt-1 Pa-1][out] drho_dt_dp: Partial derivative of alpha with respect to pressure [kg m-3 degC-1 Pa-1][in] start: Starting index within the array[in] npts: The number of values to calculateeos: Equation of state structure
-
subroutine
-
interface
calculate_density_second_derivs_linear¶ For a given thermodynamic state, return the second derivatives of density with various combinations of temperature, salinity, and pressure. Note that with a simple linear equation of state these second derivatives are all 0.
Private Functions
-
subroutine
calculate_density_second_derivs_scalar_linear(T T, S S, pressure pressure, drho_dS_dS drho_dS_dS, drho_dS_dT drho_dS_dT, drho_dT_dT drho_dT_dT, drho_dS_dP drho_dS_dP, drho_dT_dP drho_dT_dP)¶ This subroutine calculates the five, partial second derivatives of density w.r.t. potential temperature and salinity and pressure which for a linear equation of state should all be 0.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: pressure [Pa].[out] drho_ds_ds: The second derivative of density with salinity [kg m-3 PSU-2].[out] drho_ds_dt: The second derivative of density with temperature and salinity [kg m-3 ppt-1 degC-1].[out] drho_dt_dt: The second derivative of density with temperature [kg m-3 degC-2].[out] drho_ds_dp: The second derivative of density with salinity and pressure [kg m-3 PSU-1 Pa-1].[out] drho_dt_dp: The second derivative of density with temperature and pressure [kg m-3 degC-1 Pa-1].
-
subroutine
calculate_density_second_derivs_array_linear(T T, S S, pressure pressure, drho_dS_dS drho_dS_dS, drho_dS_dT drho_dS_dT, drho_dT_dT drho_dT_dT, drho_dS_dP drho_dS_dP, drho_dT_dP drho_dT_dP, start start, npts npts)¶ This subroutine calculates the five, partial second derivatives of density w.r.t. potential temperature and salinity and pressure which for a linear equation of state should all be 0.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: pressure [Pa].[out] drho_ds_ds: The second derivative of density with salinity [kg m-3 PSU-2].[out] drho_ds_dt: The second derivative of density with temperature and salinity [kg m-3 ppt-1 degC-1].[out] drho_dt_dt: The second derivative of density with temperature [kg m-3 degC-2].[out] drho_ds_dp: The second derivative of density with salinity and pressure [kg m-3 PSU-1 Pa-1].[out] drho_dt_dp: The second derivative of density with temperature and pressure [kg m-3 degC-1 Pa-1].[in] start: The starting point in the arrays.[in] npts: The number of values to calculate.
-
subroutine
-
interface
calculate_density_second_derivs_teos10¶ For a given thermodynamic state, return the second derivatives of density with various combinations of conservative temperature, absolute salinity, and pressure, using the TEOS10 expressions.
Private Functions
-
subroutine
calculate_density_second_derivs_scalar_teos10(T T, S S, pressure pressure, drho_dS_dS drho_dS_dS, drho_dS_dT drho_dS_dT, drho_dT_dT drho_dT_dT, drho_dS_dP drho_dS_dP, drho_dT_dP drho_dT_dP)¶ Calculate the 5 second derivatives of the equation of state for scalar inputs.
- Parameters
[in] t: Conservative temperature [degC][in] s: Absolute Salinity [g kg-1][in] pressure: pressure [Pa].[out] drho_ds_ds: Partial derivative of beta with respect to S[out] drho_ds_dt: Partial derivative of beta with resepct to T[out] drho_dt_dt: Partial derivative of alpha with respect to T[out] drho_ds_dp: Partial derivative of beta with respect to pressure[out] drho_dt_dp: Partial derivative of alpha with respect to pressure
-
subroutine
calculate_density_second_derivs_array_teos10(T T, S S, pressure pressure, drho_dS_dS drho_dS_dS, drho_dS_dT drho_dS_dT, drho_dT_dT drho_dT_dT, drho_dS_dP drho_dS_dP, drho_dT_dP drho_dT_dP, start start, npts npts)¶ Calculate the 5 second derivatives of the equation of state for scalar inputs.
- Parameters
[in] t: Conservative temperature [degC][in] s: Absolute Salinity [g kg-1][in] pressure: pressure [Pa].[out] drho_ds_ds: Partial derivative of beta with respect to S[out] drho_ds_dt: Partial derivative of beta with resepct to T[out] drho_dt_dt: Partial derivative of alpha with respect to T[out] drho_ds_dp: Partial derivative of beta with respect to pressure[out] drho_dt_dp: Partial derivative of alpha with respect to pressure[in] start: The starting point in the arrays.[in] npts: The number of values to calculate.
-
subroutine
-
interface
calculate_density_second_derivs_wright¶ For a given thermodynamic state, return the second derivatives of density with various combinations of temperature, salinity, and pressure.
Private Functions
-
subroutine
calculate_density_second_derivs_scalar_wright(T T, S S, P P, drho_ds_ds drho_ds_ds, drho_ds_dt drho_ds_dt, drho_dt_dt drho_dt_dt, drho_ds_dp drho_ds_dp, drho_dt_dp drho_dt_dp)¶ Second derivatives of density with respect to temperature, salinity, and pressure for scalar inputs. Inputs promoted to 1-element array and output demoted to scalar.
- Parameters
[in] t: Potential temperature referenced to 0 dbar[in] s: Salinity [PSU][in] p: pressure [Pa][out] drho_ds_ds: Partial derivative of beta with respect to S [kg m-3 PSU-2][out] drho_ds_dt: Partial derivative of beta with respcct to T [kg m-3 PSU-1 degC-1][out] drho_dt_dt: Partial derivative of alpha with respect to T [kg m-3 degC-2][out] drho_ds_dp: Partial derivative of beta with respect to pressure [kg m-3 PSU-1 Pa-1][out] drho_dt_dp: Partial derivative of alpha with respect to pressure [kg m-3 degC-1 Pa-1]
-
subroutine
calculate_density_second_derivs_array_wright(T T, S S, P P, drho_ds_ds drho_ds_ds, drho_ds_dt drho_ds_dt, drho_dt_dt drho_dt_dt, drho_ds_dp drho_ds_dp, drho_dt_dp drho_dt_dp, start start, npts npts)¶ Second derivatives of density with respect to temperature, salinity, and pressure.
- Parameters
[in] t: Potential temperature referenced to 0 dbar [degC][in] s: Salinity [PSU][in] p: Pressure [Pa][out] drho_ds_ds: Partial derivative of beta with respect to S [kg m-3 PSU-2][out] drho_ds_dt: Partial derivative of beta with respcct to T [kg m-3 PSU-1 degC-1][out] drho_dt_dt: Partial derivative of alpha with respect to T [kg m-3 degC-2][out] drho_ds_dp: Partial derivative of beta with respect to pressure [kg m-3 PSU-1 Pa-1][out] drho_dt_dp: Partial derivative of alpha with respect to pressure [kg m-3 degC-1 Pa-1][in] start: Starting index in T,S,P[in] npts: Number of points to loop over
-
subroutine
-
interface
calculate_density_teos10¶ Compute the in situ density of sea water ([kg m-3]), or its anomaly with respect to a reference density, from absolute salinity (g/kg), conservative temperature (in deg C), and pressure [Pa], using the TEOS10 expressions.
Private Functions
-
subroutine
calculate_density_scalar_teos10(T T, S S, pressure pressure, rho rho, rho_ref rho_ref)¶ This subroutine computes the in situ density of sea water (rho in [kg m-3]) from absolute salinity (S [g kg-1]), conservative temperature (T [degC]), and pressure [Pa]. It uses the expression from the TEOS10 website.
- Parameters
[in] t: Conservative temperature [degC].[in] s: Absolute salinity [g kg-1].[in] pressure: pressure [Pa].[out] rho: In situ density [kg m-3].[in] rho_ref: A reference density [kg m-3].
-
subroutine
calculate_density_array_teos10(T T, S S, pressure pressure, rho rho, start start, npts npts, rho_ref rho_ref)¶ This subroutine computes the in situ density of sea water (rho in [kg m-3]) from absolute salinity (S [g kg-1]), conservative temperature (T [degC]), and pressure [Pa]. It uses the expression from the TEOS10 website.
- Parameters
[in] t: Conservative temperature [degC].[in] s: Absolute salinity [g kg-1][in] pressure: pressure [Pa].[out] rho: in situ density [kg m-3].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.[in] rho_ref: A reference density [kg m-3].
-
subroutine
-
interface
calculate_density_unesco¶ Compute the in situ density of sea water (in [kg m-3]), or its anomaly with respect to a reference density, from salinity [PSU], potential temperature [degC], and pressure [Pa], using the UNESCO (1981) equation of state.
Private Functions
-
subroutine
calculate_density_scalar_unesco(T T, S S, pressure pressure, rho rho, rho_ref rho_ref)¶ This subroutine computes the in situ density of sea water (rho in [kg m-3]) from salinity (S [PSU]), potential temperature (T [degC]), and pressure [Pa], using the UNESCO (1981) equation of state.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: pressure [Pa].[out] rho: In situ density [kg m-3].[in] rho_ref: A reference density [kg m-3].
-
subroutine
calculate_density_array_unesco(T T, S S, pressure pressure, rho rho, start start, npts npts, rho_ref rho_ref)¶ This subroutine computes the in situ density of sea water (rho in [kg m-3]) from salinity (S [PSU]), potential temperature (T [degC]), and pressure [Pa], using the UNESCO (1981) equation of state.
- Parameters
[in] t: potential temperature relative to the surface [degC].[in] s: salinity [PSU].[in] pressure: pressure [Pa].[out] rho: in situ density [kg m-3].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.[in] rho_ref: A reference density [kg m-3].
-
subroutine
-
interface
calculate_density_wright¶ Compute the in situ density of sea water (in [kg m-3]), or its anomaly with respect to a reference density, from salinity (in psu), potential temperature (in deg C), and pressure [Pa], using the expressions from Wright, 1997, J. Atmos. Ocean. Tech., 14, 735-740.
Private Functions
-
subroutine
calculate_density_scalar_wright(T T, S S, pressure pressure, rho rho, rho_ref rho_ref)¶ This subroutine computes the in situ density of sea water (rho in [kg m-3]) from salinity (S [PSU]), potential temperature (T [degC]), and pressure [Pa]. It uses the expression from Wright, 1997, J. Atmos. Ocean. Tech., 14, 735-740.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: pressure [Pa].[out] rho: In situ density [kg m-3].[in] rho_ref: A reference density [kg m-3].
-
subroutine
calculate_density_array_wright(T T, S S, pressure pressure, rho rho, start start, npts npts, rho_ref rho_ref)¶ This subroutine computes the in situ density of sea water (rho in [kg m-3]) from salinity (S [PSU]), potential temperature (T [degC]), and pressure [Pa]. It uses the expression from Wright, 1997, J. Atmos. Ocean. Tech., 14, 735-740.
- Parameters
[in] t: potential temperature relative to the surface [degC].[in] s: salinity [PSU].[in] pressure: pressure [Pa].[out] rho: in situ density [kg m-3].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.[in] rho_ref: A reference density [kg m-3].
-
subroutine
-
interface
calculate_spec_vol¶ Calculates specific volume of sea water from T, S and P.
Private Functions
-
subroutine
calculate_spec_vol_scalar(T T, S S, pressure pressure, specvol specvol, EOS EOS, spv_ref spv_ref)¶ Calls the appropriate subroutine to calculate specific volume of sea water for scalar inputs.
- Parameters
[in] t: Potential temperature referenced to the surface [degC][in] s: Salinity [ppt][in] pressure: Pressure [Pa][out] specvol: specific volume (in-situ if pressure is local) [m3 kg-1]eos: Equation of state structure[in] spv_ref: A reference specific volume [m3 kg-1].
-
subroutine
calculate_spec_vol_array(T T, S S, pressure pressure, specvol specvol, start start, npts npts, EOS EOS, spv_ref spv_ref)¶ Calls the appropriate subroutine to calculate the specific volume of sea water for 1-D array inputs.
- Parameters
[in] t: potential temperature relative to the surface [degC].[in] s: salinity [ppt].[in] pressure: pressure [Pa].[out] specvol: in situ specific volume [kg m-3].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.eos: Equation of state structure[in] spv_ref: A reference specific volume [m3 kg-1].
-
subroutine
-
interface
calculate_spec_vol_linear¶ Compute the specific volume of sea water (in m^3/kg), or its anomaly from a reference value, using a simple linear equation of state from salinity (in psu), potential temperature (in deg C) and pressure [Pa].
Private Functions
-
subroutine
calculate_spec_vol_scalar_linear(T T, S S, pressure pressure, specvol specvol, Rho_T0_S0 Rho_T0_S0, dRho_dT dRho_dT, dRho_dS dRho_dS, spv_ref spv_ref)¶ This subroutine computes the in situ specific volume of sea water (specvol in [m3 kg-1]) from salinity (S [PSU]), potential temperature (T [degC]) and pressure [Pa], using a trivial linear equation of state for density. If spv_ref is present, specvol is an anomaly from spv_ref.
- Parameters
[in] t: potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: Pressure [Pa].[out] specvol: In situ specific volume [m3 kg-1].[in] rho_t0_s0: The density at T=0, S=0 [kg m-3].[in] drho_dt: The derivatives of density with temperature [kg m-3 degC-1].[in] drho_ds: The derivatives of density with salinity [kg m-3 ppt-1].[in] spv_ref: A reference specific volume [m3 kg-1].
-
subroutine
calculate_spec_vol_array_linear(T T, S S, pressure pressure, specvol specvol, start start, npts npts, Rho_T0_S0 Rho_T0_S0, dRho_dT dRho_dT, dRho_dS dRho_dS, spv_ref spv_ref)¶ This subroutine computes the in situ specific volume of sea water (specvol in [m3 kg-1]) from salinity (S [PSU]), potential temperature (T [degC]) and pressure [Pa], using a trivial linear equation of state for density. If spv_ref is present, specvol is an anomaly from spv_ref.
- Parameters
[in] t: potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: Pressure [Pa].[out] specvol: in situ specific volume [m3 kg-1].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.[in] rho_t0_s0: The density at T=0, S=0 [kg m-3].[in] drho_dt: The derivatives of density with temperature [kg m-3 degC-1].[in] drho_ds: The derivatives of density with salinity [kg m-3 ppt-1].[in] spv_ref: A reference specific volume [m3 kg-1].
-
subroutine
-
interface
calculate_spec_vol_teos10¶ Compute the in situ specific volume of sea water (in [m3 kg-1]), or an anomaly with respect to a reference specific volume, from absolute salinity (in g/kg), conservative temperature (in deg C), and pressure [Pa], using the TEOS10 expressions.
Private Functions
-
subroutine
calculate_spec_vol_scalar_teos10(T T, S S, pressure pressure, specvol specvol, spv_ref spv_ref)¶ This subroutine computes the in situ specific volume of sea water (specvol in [m3 kg-1]) from absolute salinity (S [g kg-1]), conservative temperature (T [degC]) and pressure [Pa], using the TEOS10 equation of state. If spv_ref is present, specvol is an anomaly from spv_ref.
- Parameters
[in] t: Conservative temperature [degC].[in] s: Absolute salinity [g kg-1][in] pressure: pressure [Pa].[out] specvol: in situ specific volume [m3 kg-1].[in] spv_ref: A reference specific volume [m3 kg-1].
-
subroutine
calculate_spec_vol_array_teos10(T T, S S, pressure pressure, specvol specvol, start start, npts npts, spv_ref spv_ref)¶ This subroutine computes the in situ specific volume of sea water (specvol in [m3 kg-1]) from absolute salinity (S [g kg-1]), conservative temperature (T [degC]) and pressure [Pa], using the TEOS10 equation of state. If spv_ref is present, specvol is an anomaly from spv_ref.
- Parameters
[in] t: Conservative temperature relative to the surface [degC].[in] s: salinity [g kg-1].[in] pressure: pressure [Pa].[out] specvol: in situ specific volume [m3 kg-1].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.[in] spv_ref: A reference specific volume [m3 kg-1].
-
subroutine
-
interface
calculate_spec_vol_unesco¶ Compute the in situ specific volume of sea water (in [m3 kg-1]), or an anomaly with respect to a reference specific volume, from salinity [PSU], potential temperature [degC], and pressure [Pa], using the UNESCO (1981) equation of state.
Private Functions
-
subroutine
calculate_spec_vol_scalar_unesco(T T, S S, pressure pressure, specvol specvol, spv_ref spv_ref)¶ This subroutine computes the in situ specific volume of sea water (specvol in [m3 kg-1]) from salinity (S [PSU]), potential temperature (T [degC]) and pressure [Pa], using the UNESCO (1981) equation of state. If spv_ref is present, specvol is an anomaly from spv_ref.
- Parameters
[in] t: potential temperature relative to the surface [degC].[in] s: salinity [PSU].[in] pressure: pressure [Pa].[out] specvol: in situ specific volume [m3 kg-1].[in] spv_ref: A reference specific volume [m3 kg-1].
-
subroutine
calculate_spec_vol_array_unesco(T T, S S, pressure pressure, specvol specvol, start start, npts npts, spv_ref spv_ref)¶ This subroutine computes the in situ specific volume of sea water (specvol in [m3 kg-1]) from salinity (S [PSU]), potential temperature (T [degC]) and pressure [Pa], using the UNESCO (1981) equation of state. If spv_ref is present, specvol is an anomaly from spv_ref.
- Parameters
[in] t: potential temperature relative to the surface [degC].[in] s: salinity [PSU].[in] pressure: pressure [Pa].[out] specvol: in situ specific volume [m3 kg-1].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.[in] spv_ref: A reference specific volume [m3 kg-1].
-
subroutine
-
interface
calculate_spec_vol_wright¶ Compute the in situ specific volume of sea water (in [m3 kg-1]), or an anomaly with respect to a reference specific volume, from salinity (in psu), potential temperature (in deg C), and pressure [Pa], using the expressions from Wright, 1997, J. Atmos. Ocean. Tech., 14, 735-740.
Private Functions
-
subroutine
calculate_spec_vol_scalar_wright(T T, S S, pressure pressure, specvol specvol, spv_ref spv_ref)¶ This subroutine computes the in situ specific volume of sea water (specvol in [m3 kg-1]) from salinity (S [PSU]), potential temperature (T [degC]) and pressure [Pa]. It uses the expression from Wright, 1997, J. Atmos. Ocean. Tech., 14, 735-740. If spv_ref is present, specvol is an anomaly from spv_ref.
- Parameters
[in] t: potential temperature relative to the surface [degC].[in] s: salinity [PSU].[in] pressure: pressure [Pa].[out] specvol: in situ specific volume [m3 kg-1].[in] spv_ref: A reference specific volume [m3 kg-1].
-
subroutine
calculate_spec_vol_array_wright(T T, S S, pressure pressure, specvol specvol, start start, npts npts, spv_ref spv_ref)¶ This subroutine computes the in situ specific volume of sea water (specvol in [m3 kg-1]) from salinity (S [PSU]), potential temperature (T [degC]) and pressure [Pa]. It uses the expression from Wright, 1997, J. Atmos. Ocean. Tech., 14, 735-740. If spv_ref is present, specvol is an anomaly from spv_ref.
- Parameters
[in] t: potential temperature relative to the surface [degC].[in] s: salinity [PSU].[in] pressure: pressure [Pa].[out] specvol: in situ specific volume [m3 kg-1].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.[in] spv_ref: A reference specific volume [m3 kg-1].
-
subroutine
-
interface
calculate_tfreeze¶ Calculates the freezing point of sea water from T, S and P.
Private Functions
-
subroutine
calculate_tfreeze_scalar(S S, pressure pressure, T_fr T_fr, EOS EOS)¶ Calls the appropriate subroutine to calculate the freezing point for scalar inputs.
- Parameters
[in] s: Salinity [ppt][in] pressure: Pressure [Pa][out] t_fr: Freezing point potential temperature referenced to the surface [degC]eos: Equation of state structure
-
subroutine
calculate_tfreeze_array(S S, pressure pressure, T_fr T_fr, start start, npts npts, EOS EOS)¶ Calls the appropriate subroutine to calculate the freezing point for a 1-D array.
- Parameters
[in] s: Salinity [ppt][in] pressure: Pressure [Pa][out] t_fr: Freezing point potential temperature referenced to the surface [degC][in] start: Starting index within the array[in] npts: The number of values to calculateeos: Equation of state structure
-
subroutine
-
interface
calculate_tfreeze_linear¶ Compute the freezing point potential temperature [degC] from salinity [ppt] and pressure [Pa] using a simple linear expression, with coefficients passed in as arguments.
Private Functions
-
subroutine
calculate_tfreeze_linear_scalar(S S, pres pres, T_Fr T_Fr, TFr_S0_P0 TFr_S0_P0, dTFr_dS dTFr_dS, dTFr_dp dTFr_dp)¶ This subroutine computes the freezing point potential temperature [degC] from salinity [ppt], and pressure [Pa] using a simple linear expression, with coefficients passed in as arguments.
- Parameters
[in] s: salinity [ppt].[in] pres: pressure [Pa].[out] t_fr: Freezing point potential temperature [degC].[in] tfr_s0_p0: The freezing point at S=0, p=0 [degC].[in] dtfr_ds: The derivative of freezing point with salinity, [degC ppt-1].[in] dtfr_dp: The derivative of freezing point with pressure, [degC Pa-1].
-
subroutine
calculate_tfreeze_linear_array(S S, pres pres, T_Fr T_Fr, start start, npts npts, TFr_S0_P0 TFr_S0_P0, dTFr_dS dTFr_dS, dTFr_dp dTFr_dp)¶ This subroutine computes an array of freezing point potential temperatures [degC] from salinity [ppt], and pressure [Pa] using a simple linear expression, with coefficients passed in as arguments.
- Parameters
[in] s: salinity [ppt].[in] pres: pressure [Pa].[out] t_fr: Freezing point potential temperature [degC].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.[in] tfr_s0_p0: The freezing point at S=0, p=0, [degC].[in] dtfr_ds: The derivative of freezing point with salinity, [degC PSU-1].[in] dtfr_dp: The derivative of freezing point with pressure, [degC Pa-1].
-
subroutine
-
interface
calculate_tfreeze_millero¶ Compute the freezing point potential temperature [degC] from salinity [PSU] and pressure [Pa] using the expression from Millero (1978) (and in appendix A of Gill 1982), but with the of the pressure dependence changed from 7.53e-8 to 7.75e-8 to make this an expression for potential temperature (not in situ temperature), using a value that is correct at the freezing point at 35 PSU and 5e6 Pa (500 dbar).
Private Functions
-
subroutine
calculate_tfreeze_millero_scalar(S S, pres pres, T_Fr T_Fr)¶ This subroutine computes the freezing point potential temperature [degC] from salinity [ppt], and pressure [Pa] using the expression from Millero (1978) (and in appendix A of Gill 1982), but with the of the pressure dependence changed from 7.53e-8 to 7.75e-8 to make this an expression for potential temperature (not in situ temperature), using a value that is correct at the freezing point at 35 PSU and 5e6 Pa (500 dbar).
- Parameters
[in] s: Salinity in PSU.[in] pres: Pressure [Pa].[out] t_fr: Freezing point potential temperature [degC].
-
subroutine
calculate_tfreeze_millero_array(S S, pres pres, T_Fr T_Fr, start start, npts npts)¶ This subroutine computes the freezing point potential temperature [degC] from salinity [ppt], and pressure [Pa] using the expression from Millero (1978) (and in appendix A of Gill 1982), but with the of the pressure dependence changed from 7.53e-8 to 7.75e-8 to make this an expression for potential temperature (not in situ temperature), using a value that is correct at the freezing point at 35 PSU and 5e6 Pa (500 dbar).
- Parameters
[in] s: Salinity [PSU].[in] pres: Pressure [Pa].[out] t_fr: Freezing point potential temperature [degC].[in] start: The starting point in the arrays.[in] npts: The number of values to calculate.
-
subroutine
-
interface
calculate_tfreeze_teos10¶ Compute the freezing point conservative temperature [degC] from absolute salinity [g/kg] and pressure [Pa] using the TEOS10 package.
Private Functions
-
subroutine
calculate_tfreeze_teos10_scalar(S S, pres pres, T_Fr T_Fr)¶ This subroutine computes the freezing point conservative temperature [degC] from absolute salinity [g/kg], and pressure [Pa] using the TEOS10 package.
- Parameters
[in] s: Absolute salinity [g/kg].[in] pres: Pressure [Pa].[out] t_fr: Freezing point conservative temperature [degC].
-
subroutine
calculate_tfreeze_teos10_array(S S, pres pres, T_Fr T_Fr, start start, npts npts)¶ This subroutine computes the freezing point conservative temperature [degC] from absolute salinity [g/kg], and pressure [Pa] using the TEOS10 package.
- Parameters
[in] s: absolute salinity [g/kg].[in] pres: pressure [Pa].[out] t_fr: Freezing point conservative temperature [degC].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.
-
subroutine
-
interface
check_redundant¶ Check for consistency between the duplicated points of a C-grid vector.
Private Functions
-
subroutine
check_redundant_vc3d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, is is, ie ie, js js, je je, direction direction)¶ Check for consistency between the duplicated points of a 3-D C-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector to be checked for consistency[in] v_comp: The u-component of the vector to be checked for consistency[in] is: The starting i-index to check[in] ie: The ending i-index to check[in] js: The starting j-index to check[in] je: The ending j-index to check[in] direction: the direction flag to be passed to pass_vector
-
subroutine
check_redundant_vc2d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, is is, ie ie, js js, je je, direction direction)¶ Check for consistency between the duplicated points of a 2-D C-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector to be checked for consistency[in] v_comp: The u-component of the vector to be checked for consistency[in] is: The starting i-index to check[in] ie: The ending i-index to check[in] js: The starting j-index to check[in] je: The ending j-index to check[in] direction: the direction flag to be passed to pass_vector
-
subroutine
-
interface
check_redundant_b¶ Check for consistency between the duplicated points of a B-grid vector or scalar.
Private Functions
-
subroutine
check_redundant_vb3d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, is is, ie ie, js js, je je, direction direction)¶ Check for consistency between the duplicated points of a 3-D B-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector to be checked for consistency[in] v_comp: The v-component of the vector to be checked for consistency[in] is: The starting i-index to check[in] ie: The ending i-index to check[in] js: The starting j-index to check[in] je: The ending j-index to check[in] direction: the direction flag to be passed to pass_vector
-
subroutine
check_redundant_vb2d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, is is, ie ie, js js, je je, direction direction)¶ Check for consistency between the duplicated points of a 2-D B-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector to be checked for consistency[in] v_comp: The v-component of the vector to be checked for consistency[in] is: The starting i-index to check[in] ie: The ending i-index to check[in] js: The starting j-index to check[in] je: The ending j-index to check[in] direction: the direction flag to be passed to pass_vector
-
subroutine
check_redundant_sb3d(mesg mesg, array array, G G, is is, ie ie, js js, je je)¶ Check for consistency between the duplicated points of a 3-D scalar at corner points.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] array: The array to be checked for consistency[in] is: The starting i-index to check[in] ie: The ending i-index to check[in] js: The starting j-index to check[in] je: The ending j-index to check
-
subroutine
check_redundant_sb2d(mesg mesg, array array, G G, is is, ie ie, js js, je je)¶ Check for consistency between the duplicated points of a 2-D scalar at corner points.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] array: The array to be checked for consistency[in] is: The starting i-index to check[in] ie: The ending i-index to check[in] js: The starting j-index to check[in] je: The ending j-index to check
-
subroutine
-
interface
check_redundant_c¶ Check for consistency between the duplicated points of a C-grid vector.
Private Functions
-
subroutine
check_redundant_vc3d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, is is, ie ie, js js, je je, direction direction)¶ Check for consistency between the duplicated points of a 3-D C-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector to be checked for consistency[in] v_comp: The u-component of the vector to be checked for consistency[in] is: The starting i-index to check[in] ie: The ending i-index to check[in] js: The starting j-index to check[in] je: The ending j-index to check[in] direction: the direction flag to be passed to pass_vector
-
subroutine
check_redundant_vc2d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, is is, ie ie, js js, je je, direction direction)¶ Check for consistency between the duplicated points of a 2-D C-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector to be checked for consistency[in] v_comp: The u-component of the vector to be checked for consistency[in] is: The starting i-index to check[in] ie: The ending i-index to check[in] js: The starting j-index to check[in] je: The ending j-index to check[in] direction: the direction flag to be passed to pass_vector
-
subroutine
-
interface
check_redundant_t¶ Check for consistency between the duplicated points of an A-grid vector or scalar.
Private Functions
-
subroutine
check_redundant_st3d(mesg mesg, array array, G G, is is, ie ie, js js, je je)¶ Check for consistency between the duplicated points of a 3-D scalar at tracer points.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] array: The array to be checked for consistency[in] is: The starting i-index to check[in] ie: The ending i-index to check[in] js: The starting j-index to check[in] je: The ending j-index to check
-
subroutine
check_redundant_st2d(mesg mesg, array array, G G, is is, ie ie, js js, je je)¶ Check for consistency between the duplicated points of a 2-D scalar at tracer points.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] array: The array to be checked for consistency[in] is: The starting i-index to check[in] ie: The ending i-index to check[in] js: The starting j-index to check[in] je: The ending j-index to check
-
subroutine
check_redundant_vt3d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, is is, ie ie, js js, je je, direction direction)¶ Check for consistency between the duplicated points of a 3-D A-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector to be checked for consistency[in] v_comp: The v-component of the vector to be checked for consistency[in] is: The starting i-index to check[in] ie: The ending i-index to check[in] js: The starting j-index to check[in] je: The ending j-index to check[in] direction: the direction flag to be passed to pass_vector
-
subroutine
check_redundant_vt2d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, is is, ie ie, js js, je je, direction direction)¶ Check for consistency between the duplicated points of a 2-D A-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector to be checked for consistency[in] v_comp: The v-component of the vector to be checked for consistency[in] is: The starting i-index to check[in] ie: The ending i-index to check[in] js: The starting j-index to check[in] je: The ending j-index to check[in] direction: the direction flag to be passed to pass_vector
-
subroutine
-
interface
chk_sum_msg¶ Write a message with either checksums or numerical statistics of arrays.
Private Functions
-
subroutine
chk_sum_msg1(fmsg fmsg, bc0 bc0, mesg mesg, iounit iounit)¶ Write a message including the checksum of the non-shifted array.
- Parameters
[in] fmsg: A checksum code-location specific preamble[in] mesg: An identifying message supplied by top-level caller[in] bc0: The bitcount of the non-shifted array[in] iounit: Checksum logger IO unit
-
subroutine
chk_sum_msg2(fmsg fmsg, bc0 bc0, bcSW bcSW, mesg mesg, iounit iounit)¶ Write a message including checksums of non-shifted and southwestward shifted arrays.
- Parameters
[in] fmsg: A checksum code-location specific preamble[in] mesg: An identifying message supplied by top-level caller[in] bc0: The bitcount of the non-shifted array[in] bcsw: The bitcount of the southwest-shifted array[in] iounit: Checksum logger IO unit
-
subroutine
chk_sum_msg3(fmsg fmsg, aMean aMean, aMin aMin, aMax aMax, mesg mesg, iounit iounit)¶ Write a message including the global mean, maximum and minimum of an array.
- Parameters
[in] fmsg: A checksum code-location specific preamble[in] mesg: An identifying message supplied by top-level caller[in] amean: The mean value of the array[in] amin: The minimum value of the array[in] amax: The maximum value of the array[in] iounit: Checksum logger IO unit
-
subroutine
chk_sum_msg5(fmsg fmsg, bc0 bc0, bcSW bcSW, bcSE bcSE, bcNW bcNW, bcNE bcNE, mesg mesg, iounit iounit)¶ Write a message including checksums of non-shifted and diagonally shifted arrays.
- Parameters
[in] fmsg: A checksum code-location specific preamble[in] mesg: An identifying message supplied by top-level caller[in] bc0: The bitcount of the non-shifted array[in] bcsw: The bitcount for SW shifted array[in] bcse: The bitcount for SE shifted array[in] bcnw: The bitcount for NW shifted array[in] bcne: The bitcount for NE shifted array[in] iounit: Checksum logger IO unit
-
subroutine
-
interface
chksum¶ This is an older interface for 1-, 2-, or 3-D checksums.
Private Functions
-
subroutine
chksum1d(array array, mesg mesg, start_i start_i, end_i end_i, compare_PEs compare_PEs)¶ chksum1d does a checksum of a 1-dimensional array.
- Parameters
[in] array: The array to be summed (index starts at 1).[in] mesg: An identifying message.[in] start_i: The starting index for the sum (default 1)[in] end_i: The ending index for the sum (default all)[in] compare_pes: If true, compare across PEs instead of summing and list the root_PE value (default true)
-
subroutine
-
interface
clone_mom_domain¶ Copy one MOM_domain_type into another.
Private Functions
-
subroutine
clone_md_to_md(MD_in MD_in, MOM_dom MOM_dom, min_halo min_halo, halo_size halo_size, symmetric symmetric, domain_name domain_name)¶ clone_MD_to_MD copies one MOM_domain_type into another, while allowing some properties of the new type to differ from the original one.
- Parameters
[in] md_in: An existing MOM_domainmom_dom: A pointer to a MOM_domain that will be allocated if it is unassociated, and will have data copied from MD_in[inout] min_halo: If present, this sets the[in] halo_size: If present, this sets the halo size for the domian in the i- and j-directions. min_halo and halo_size can not both be present.[in] symmetric: If present, this specifies whether the new domain is symmetric, regardless of whether the macro SYMMETRIC_MEMORY_ is defined.[in] domain_name: A name for the new domain, “MOM”
-
subroutine
clone_md_to_d2d(MD_in MD_in, mpp_domain mpp_domain, min_halo min_halo, halo_size halo_size, symmetric symmetric, domain_name domain_name)¶ clone_MD_to_d2D uses information from a MOM_domain_type to create a new domain2d type, while allowing some properties of the new type to differ from the original one.
- Parameters
[in] md_in: An existing MOM_domain to be cloned[inout] mpp_domain: The new mpp_domain to be set up[inout] min_halo: If present, this sets the[in] halo_size: If present, this sets the halo size for the domian in the i- and j-directions. min_halo and halo_size can not both be present.[in] symmetric: If present, this specifies whether the new domain is symmetric, regardless of whether the macro SYMMETRIC_MEMORY_ is defined.[in] domain_name: A name for the new domain, “MOM”
-
subroutine
- type
Pointers to arrays with transports, which can later be used for derived diagnostics, like energy balances.
Public Members
-
real, dimension(:,:,:), pointer mom_variables::cont_diag_ptrs::uh => NULL() Resolved zonal layer thickness fluxes, [H m2 s-1 ~> m3 s-1 or kg s-1].
-
real, dimension(:,:,:), pointer mom_variables::cont_diag_ptrs::vh => NULL() Resolved meridional layer thickness fluxes, [H m2 s-1 ~> m3 s-1 or kg s-1].
-
real, dimension(:,:,:), pointer mom_variables::cont_diag_ptrs::uhgm => NULL() Isopycnal height diffusion induced zonal volume fluxes [H m2 s-1 ~> m3 s-1 or kg s-1].
-
real, dimension(:,:,:), pointer mom_variables::cont_diag_ptrs::vhgm => NULL() Isopycnal height diffusion induced meridional volume fluxes [H m2 s-1 ~> m3 s-1 or kg s-1].
-
real, dimension(:,:,:), pointer mom_variables::cont_diag_ptrs::diapyc_vel => NULL() The net diapycnal velocity [H s-1 ~> m s-1 or kg m-2 s-1].
-
- type
Control structure for mom_continuity.
Public Members
-
integer
continuity_scheme¶ Selects the discretization for the continuity solver. Valid values are:
PPM - A directionally split piecewise parabolic reconstruction solver. The default, PPM, seems most appropriate for use with our current time-splitting strategies.
-
type(continuity_ppm_cs), pointer mom_continuity::continuity_cs::ppm_csp => NULL() Control structure for mom_continuity_ppm.
-
integer
- type
Control structure for mom_continuity_ppm.
Unnamed Group
-
type(diag_ctrl), pointer mom_continuity_ppm::continuity_ppm_cs::diag Diagnostics control structure.
-
logical
upwind_1st¶ If true, use a first-order upwind scheme.
-
logical
monotonic¶ If true, use the Colella & Woodward monotonic limiter; otherwise use a simple positive definite limiter.
-
logical
simple_2nd¶ If true, use a simple second order (arithmetic mean) interpolation of the edge values instead of the higher order interpolation.
-
real
tol_eta¶ The tolerance for free-surface height discrepancies between the barotropic solution and the sum of the layer thicknesses [H ~> m or kg m-2].
-
real
tol_vel¶ The tolerance for barotropic velocity discrepancies between the barotropic solution and the sum of the layer thicknesses [m s-1].
-
real
tol_eta_aux¶ The tolerance for free-surface height discrepancies between the barotropic solution and the sum of the layer thicknesses when calculating the auxiliary corrected velocities [H ~> m or kg m-2].
-
real
cfl_limit_adjust¶ The maximum CFL of the adjusted velocities [nondim].
-
logical
aggress_adjust¶ If true, allow the adjusted velocities to have a relative CFL change up to 0.5. False by default.
-
logical
vol_cfl¶ If true, use the ratio of the open face lengths to the tracer cell areas when estimating CFL numbers. Without aggress_adjust, the default is false; it is always true with.
-
logical
better_iter¶ If true, stop corrective iterations using a velocity-based criterion and only stop if the iteration is better than all predecessors.
-
logical
use_visc_rem_max¶ If true, use more appropriate limiting bounds for corrections in strongly viscous columns.
-
logical
marginal_faces¶ If true, use the marginal face areas from the continuity solver for use as the weights in the barotropic solver. Otherwise use the transport averaged areas.
-
-
interface
coordinateunits¶ Returns a string with the coordinate units associated with the coordinate mode.
Public Functions
-
character(len=16) function regrid_consts::coordinateunits::coordinateunitsi(coordMode coordMode) Returns a string with the coordinate units associated with the enumerated integer,.
- Return
Units of coordinate
- Parameters
[in] coordmode: Coordinate mode
-
character(len=16) function regrid_consts::coordinateunits::coordinateunitss(string string) Returns a string with the coordinate units associated with the string defining the coordinate mode.
- Return
Units of coordinate
- Parameters
[in] string: Coordinate mode
-
- type
Control structure for mom_coriolisadv.
Unnamed Group
-
integer
id_rv= -1¶ Diagnostic IDs.
-
integer
id_pv= -1¶ Diagnostic IDs.
-
integer
id_gkeu= -1¶ Diagnostic IDs.
-
integer
id_gkev= -1¶ Diagnostic IDs.
-
integer
id_rvxu= -1¶ Diagnostic IDs.
-
integer
id_rvxv= -1¶ Diagnostic IDs.
Public Members
-
integer
coriolis_scheme¶ Selects the discretization for the Coriolis terms. Valid values are:
SADOURNY75_ENERGY - Sadourny, 1975
ARAKAWA_HSU90 - Arakawa & Hsu, 1990, Energy & non-div. Enstrophy
ROBUST_ENSTRO - Pseudo-enstrophy scheme
SADOURNY75_ENSTRO - Sadourny, JAS 1975, Enstrophy
ARAKAWA_LAMB81 - Arakawa & Lamb, MWR 1981, Energy & Enstrophy
ARAKAWA_LAMB_BLEND - A blend of Arakawa & Lamb with Arakawa & Hsu and Sadourny energy. The default, SADOURNY75_ENERGY, is the safest choice then the deformation radius is poorly resolved.
-
integer
ke_scheme¶ KE_SCHEME selects the discretization for the kinetic energy. Valid values are: KE_ARAKAWA, KE_SIMPLE_GUDONOV, KE_GUDONOV.
-
integer
pv_adv_scheme¶ PV_ADV_SCHEME selects the discretization for PV advection Valid values are:
PV_ADV_CENTERED - centered (aka Sadourny, 75)
PV_ADV_UPWIND1 - upwind, first order
-
real
f_eff_max_blend¶ The factor by which the maximum effective Coriolis acceleration from any point can be increased when blending different discretizations with the ARAKAWA_LAMB_BLEND Coriolis scheme. This must be greater than 2.0, and is 4.0 by default.
-
real
wt_lin_blend¶ A weighting value beyond which the blending between Sadourny and Arakawa & Hsu goes linearly to 0. This must be between 1 and 1e-15, often 1/8.
-
logical
no_slip¶ If true, no slip boundary conditions are used. Otherwise free slip boundary conditions are assumed. The implementation of the free slip boundary conditions on a C-grid is much cleaner than the no slip boundary conditions. The use of free slip b.c.s is strongly encouraged. The no slip b.c.s are not implemented with the biharmonic viscosity.
-
logical
bound_coriolis¶ If true, the Coriolis terms at u points are bounded by the four estimates of (f+rv)v from the four neighboring v points, and similarly at v points. This option would have no effect on the SADOURNY75_ENERGY scheme if it were possible to use centered difference thickness fluxes.
-
logical
coriolis_en_dis¶ If CORIOLIS_EN_DIS is defined, two estimates of the thickness fluxes are used to estimate the Coriolis term, and the one that dissipates energy relative to the other one is used. This is only available at present if Coriolis scheme is SADOURNY75_ENERGY.
-
type(time_type), pointer mom_coriolisadv::coriolisadv_cs::time A pointer to the ocean model’s clock.
-
type(diag_ctrl), pointer mom_coriolisadv::coriolisadv_cs::diag A structure that is used to regulate the timing of diagnostic output.
-
integer
-
interface
create_group_pass¶ Set up a group of halo updates.
Private Functions
-
subroutine
create_var_group_pass_2d(group group, array array, MOM_dom MOM_dom, sideflag sideflag, position position, halo halo, clock clock)¶ create_var_group_pass_2d sets up a group of two-dimensional array halo updates.
- Parameters
[inout] group: The data type that store information for group update. This data will be used in do_group_pass.[inout] array: The array which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] sideflag: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if sideflag is omitted.[in] position: An optional argument indicating the position. This is usally CORNER, but is CENTER by default.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
create_var_group_pass_3d(group group, array array, MOM_dom MOM_dom, sideflag sideflag, position position, halo halo, clock clock)¶ create_var_group_pass_3d sets up a group of three-dimensional array halo updates.
- Parameters
[inout] group: The data type that store information for group update. This data will be used in do_group_pass.[inout] array: The array which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] sideflag: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if sideflag is omitted.[in] position: An optional argument indicating the position. This is usally CORNER, but is CENTER by default.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
create_vector_group_pass_2d(group group, u_cmpt u_cmpt, v_cmpt v_cmpt, MOM_dom MOM_dom, direction direction, stagger stagger, halo halo, clock clock)¶ create_vector_group_pass_2d sets up a group of two-dimensional vector halo updates.
- Parameters
[inout] group: The data type that store information for group update. This data will be used in do_group_pass.[inout] u_cmpt: The nominal zonal (u) component of the vector pair which is having its halos points exchanged.[inout] v_cmpt: The nominal meridional (v) component of the vector pair which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent[in] direction: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH, possibly plus SCALAR_PAIR if these are paired non-directional scalars discretized at the typical vector component locations. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if omitted.[in] stagger: An optional flag, which may be one of A_GRID, BGRID_NE, or CGRID_NE, indicating where the two components of the vector are discretized. Omitting stagger is the same as setting it to CGRID_NE.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
create_vector_group_pass_3d(group group, u_cmpt u_cmpt, v_cmpt v_cmpt, MOM_dom MOM_dom, direction direction, stagger stagger, halo halo, clock clock)¶ create_vector_group_pass_3d sets up a group of three-dimensional vector halo updates.
- Parameters
[inout] group: The data type that store information for group update. This data will be used in do_group_pass.[inout] u_cmpt: The nominal zonal (u) component of the vector pair which is having its halos points exchanged.[inout] v_cmpt: The nominal meridional (v) component of the vector pair which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] direction: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH, possibly plus SCALAR_PAIR if these are paired non-directional scalars discretized at the typical vector component locations. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if omitted.[in] stagger: An optional flag, which may be one of A_GRID, BGRID_NE, or CGRID_NE, indicating where the two components of the vector are discretized. Omitting stagger is the same as setting it to CGRID_NE.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
- type
Control structure for MOM_controlled_forcing.
Unnamed Group
-
real, dimension(:), pointer mom_controlled_forcing::ctrl_forcing_cs::avg_time => NULL() Pointers for data.
-
real, dimension(:,:), pointer mom_controlled_forcing::ctrl_forcing_cs::heat_0 => NULL() Pointers for data.
-
real, dimension(:,:), pointer mom_controlled_forcing::ctrl_forcing_cs::precip_0 => NULL() Pointers for data.
-
real, dimension(:,:,:), pointer mom_controlled_forcing::ctrl_forcing_cs::heat_cyc => NULL() Pointers for data.
-
real, dimension(:,:,:), pointer mom_controlled_forcing::ctrl_forcing_cs::precip_cyc => NULL() Pointers for data.
-
real, dimension(:,:,:), pointer mom_controlled_forcing::ctrl_forcing_cs::avg_sst_anom => NULL() Pointers for data.
-
real, dimension(:,:,:), pointer mom_controlled_forcing::ctrl_forcing_cs::avg_sss_anom => NULL() Pointers for data.
-
real, dimension(:,:,:), pointer mom_controlled_forcing::ctrl_forcing_cs::avg_sss => NULL() Pointers for data.
-
type(diag_ctrl), pointer mom_controlled_forcing::ctrl_forcing_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
integer
id_heat_0= -1¶ Diagnostic handle.
Public Members
-
logical
use_temperature¶ If true, temperature and salinity are used as state variables.
-
logical
do_integrated¶ If true, use time-integrated anomalies to control the surface state.
-
integer
num_cycle¶ The number of elements in the forcing cycle.
-
real
heat_int_rate¶ The rate at which heating anomalies accumulate [s-1].
-
real
prec_int_rate¶ The rate at which precipitation anomalies accumulate [s-1].
-
real
heat_cyc_rate¶ The rate at which cyclical heating anomaliess accumulate [s-1].
-
real
prec_cyc_rate¶ The rate at which cyclical precipitation anomaliess accumulate [s-1].
-
real
len2¶ The square of the length scale over which the anomalies are smoothed via a Laplacian filter [m2].
-
real
lam_heat¶ A constant of proportionality between SST anomalies and heat fluxes [W m-2 degC-1].
-
real
lam_prec¶ A constant of proportionality between SSS anomalies (normalised by mean SSS) and precipitation [kg m-2].
-
real
lam_cyc_heat¶ A constant of proportionality between cyclical SST anomalies and corrective heat fluxes [W m-2 degC-1].
-
real
lam_cyc_prec¶ A constant of proportionality between cyclical SSS anomalies (normalised by mean SSS) and corrective precipitation [kg m-2].
-
- type
Control structure including parameters for CVMix convection.
Unnamed Group
-
integer
id_n2= -1¶ Diagnostics handles.
-
integer
id_kd_conv= -1¶ Diagnostics handles.
-
integer
id_kv_conv= -1¶ Diagnostics handles.
-
real, dimension(:,:,:), allocatable mom_cvmix_conv::cvmix_conv_cs::n2 Squared Brunt-Vaisala frequency [s-2].
-
real, dimension(:,:,:), allocatable mom_cvmix_conv::cvmix_conv_cs::kd_conv Diffusivity added by convection [Z2 T-1 ~> m2 s-1].
-
real, dimension(:,:,:), allocatable mom_cvmix_conv::cvmix_conv_cs::kv_conv Viscosity added by convection [Z2 T-1 ~> m2 s-1].
Public Members
-
real
kd_conv_const¶ diffusivity constant used in convective regime [m2 s-1]
-
real
kv_conv_const¶ viscosity constant used in convective regime [m2 s-1]
-
real
bv_sqr_conv¶ Threshold for squared buoyancy frequency needed to trigger Brunt-Vaisala parameterization [s-2].
-
real
min_thickness¶ Minimum thickness allowed [m].
-
logical
debug¶ If true, turn on debugging.
-
type(diag_ctrl), pointer mom_cvmix_conv::cvmix_conv_cs::diag => NULL() Pointer to diagnostics control structure.
-
integer
- type
Control structure including parameters for CVMix double diffusion.
Unnamed Group
-
integer
id_kt_extra= -1¶ Diagnostics handles.
-
integer
id_ks_extra= -1¶ Diagnostics handles.
-
integer
id_r_rho= -1¶ Diagnostics handles.
-
real, dimension(:,:,:), allocatable mom_cvmix_ddiff::cvmix_ddiff_cs::r_rho Double-diffusion density ratio [nondim].
Public Members
-
real
strat_param_max¶ maximum value for the stratification parameter [nondim]
-
real
kappa_ddiff_s¶ leading coefficient in formula for salt-fingering regime for salinity diffusion [m2 s-1]
-
real
ddiff_exp1¶ interior exponent in salt-fingering regime formula [nondim]
-
real
ddiff_exp2¶ exterior exponent in salt-fingering regime formula [nondim]
-
real
mol_diff¶ molecular diffusivity [m2 s-1]
-
real
kappa_ddiff_param1¶ exterior coefficient in diffusive convection regime [nondim]
-
real
kappa_ddiff_param2¶ middle coefficient in diffusive convection regime [nondim]
-
real
kappa_ddiff_param3¶ interior coefficient in diffusive convection regime [nondim]
-
real
min_thickness¶ Minimum thickness allowed [m].
-
character(len=4) mom_cvmix_ddiff::cvmix_ddiff_cs::diff_conv_type type of diffusive convection to use. Options are Marmorino & Caldwell 1976 (“MC76”; default) and Kelley 1988, 1990 (“K90”)
-
logical
debug¶ If true, turn on debugging.
-
type(diag_ctrl), pointer mom_cvmix_ddiff::cvmix_ddiff_cs::diag => NULL() Pointer to diagnostics control structure.
-
integer
- type
Control structure including parameters for CVMix interior shear schemes.
Unnamed Group
-
integer
id_n2= -1¶ Diagnostic handles.
-
integer
id_s2= -1¶ Diagnostic handles.
-
integer
id_ri_grad= -1¶ Diagnostic handles.
-
integer
id_kv= -1¶ Diagnostic handles.
-
integer
id_kd= -1¶ Diagnostic handles.
-
integer
id_ri_grad_smooth= -1¶ Diagnostic handles.
Public Members
-
logical
use_lmd94¶ Flags to use the LMD94 scheme.
-
logical
use_pp81¶ Flags to use Pacanowski and Philander (JPO 1981)
-
logical
smooth_ri¶ If true, smooth Ri using a 1-2-1 filter.
-
real
ri_zero¶ LMD94 critical Richardson number.
-
real
nu_zero¶ LMD94 maximum interior diffusivity.
-
real
kpp_exp¶ Exponent of unitless factor of diff. for KPP internal shear mixing scheme.
-
real, dimension(:,:,:), allocatable mom_cvmix_shear::cvmix_shear_cs::n2 Squared Brunt-Vaisala frequency [s-2].
-
real, dimension(:,:,:), allocatable mom_cvmix_shear::cvmix_shear_cs::s2 Squared shear frequency [s-2].
-
real, dimension(:,:,:), allocatable mom_cvmix_shear::cvmix_shear_cs::ri_grad Gradient Richardson number.
-
real, dimension(:,:,:), allocatable mom_cvmix_shear::cvmix_shear_cs::ri_grad_smooth Gradient Richardson number after smoothing.
-
character(10) mom_cvmix_shear::cvmix_shear_cs::mix_scheme Mixing scheme name (string)
-
type(diag_ctrl), pointer mom_cvmix_shear::cvmix_shear_cs::diag => NULL() Pointer to the diagnostics control structure.
-
integer
- type
A list of depths and corresponding globally integrated ocean area at each depth and the ocean volume below each depth.
- type
Control structure for diabatic_aux.
Public Members
-
logical
do_rivermix= .false.¶ Provide additional TKE to mix river runoff at the river mouths to a depth of “rivermix_depth”.
-
real
rivermix_depth= 0.0¶ The depth to which rivers are mixed if do_rivermix = T [Z ~> m].
-
logical
reclaim_frazil¶ If true, try to use any frazil heat deficit to to cool the topmost layer down to the freezing point. The default is false.
-
logical
pressure_dependent_frazil¶ If true, use a pressure dependent freezing temperature when making frazil. The default is false, which will be faster but is inappropriate with ice-shelf cavities.
-
logical
ignore_fluxes_over_land¶ If true, the model does not check if fluxes are applied over land points. This flag must be used when the ocean is coupled with sea ice and ice shelves and use_ePBL = true.
-
logical
use_river_heat_content¶ If true, assumes that ice-ocean boundary has provided a river heat content. Otherwise, runoff is added with a temperature of the local SST.
-
logical
use_calving_heat_content¶ If true, assumes that ice-ocean boundary has provided a calving heat content. Otherwise, calving is added with a temperature of the local SST.
-
logical
var_pen_sw¶ If true, use one of the CHL_A schemes to determine the e-folding depth of incoming shortwave radiation.
-
integer
sbc_chl¶ An integer handle used in time interpolation of chlorophyll read from a file.
-
logical
chl_from_file¶ If true, chl_a is read from a file.
-
type(time_type), pointer mom_diabatic_aux::diabatic_aux_cs::time => NULL() A pointer to the ocean model’s clock.
-
type(diag_ctrl), pointer mom_diabatic_aux::diabatic_aux_cs::diag Structure used to regulate timing of diagnostic output.
-
integer
id_createdh= -1¶ Diagnostic ID of mass added to avoid grounding.
-
integer
id_brine_lay= -1¶ Diagnostic ID of which layer receives the brine.
-
integer
id_pensw_diag= -1¶ Diagnostic ID of Penetrative shortwave heating (flux convergence)
-
integer
id_penswflux_diag= -1¶ Diagnostic ID of Penetrative shortwave flux.
-
integer
id_nonpensw_diag= -1¶ Diagnostic ID of Non-penetrative shortwave heating.
-
integer
id_chl= -1¶ Diagnostic ID of chlorophyll-A handles for opacity.
-
real, dimension(:,:), allocatable mom_diabatic_aux::diabatic_aux_cs::createdh The amount of volume added in order to avoid grounding [m s-1].
-
real, dimension(:,:,:), allocatable mom_diabatic_aux::diabatic_aux_cs::pensw_diag Heating in a layer from convergence of penetrative SW [W m-2].
-
real, dimension(:,:,:), allocatable mom_diabatic_aux::diabatic_aux_cs::penswflux_diag Penetrative SW flux at base of grid layer [W m-2].
-
real, dimension(:,:), allocatable mom_diabatic_aux::diabatic_aux_cs::nonpensw_diag Non-downwelling SW radiation at ocean surface [W m-2].
-
logical
- type
Control structure for this module.
Unnamed Group
-
integer
id_cg1= -1¶ Diagnostic IDs.
-
integer, dimension(:), allocatable mom_diabatic_driver::diabatic_cs::id_cn Diagnostic IDs.
-
integer
id_wd= -1¶ Diagnostic IDs.
-
integer
id_ea= -1¶ Diagnostic IDs.
-
integer
id_eb= -1¶ Diagnostic IDs.
-
integer
id_dudt_dia= -1¶ Diagnostic IDs.
-
integer
id_dvdt_dia= -1¶ Diagnostic IDs.
-
integer
id_ea_s= -1¶ Diagnostic IDs.
-
integer
id_eb_s= -1¶ Diagnostic IDs.
-
integer
id_ea_t= -1¶ Diagnostic IDs.
-
integer
id_eb_t= -1¶ Diagnostic IDs.
-
integer
id_kd_heat= -1¶ Diagnostic IDs.
-
integer
id_kd_salt= -1¶ Diagnostic IDs.
-
integer
id_kd_interface= -1¶ Diagnostic IDs.
-
integer
id_kd_epbl= -1¶ Diagnostic IDs.
-
integer
id_tdif= -1¶ Diagnostic IDs.
-
integer
id_tadv= -1¶ Diagnostic IDs.
-
integer
id_sdif= -1¶ Diagnostic IDs.
-
integer
id_sadv= -1¶ Diagnostic IDs.
-
integer
id_mld_003= -1¶ Diagnostic IDs.
-
integer
id_mld_0125= -1¶ Diagnostic IDs.
-
integer
id_mld_user= -1¶ Diagnostic IDs.
-
integer
id_mlotstsq= -1¶ Diagnostic IDs.
-
integer
id_submln2= -1¶ Diagnostic IDs.
-
integer
id_brine_lay= -1¶ Diagnostic IDs.
-
integer
id_u_predia= -1¶ Diagnostic IDs.
-
integer
id_v_predia= -1¶ Diagnostic IDs.
-
integer
id_h_predia= -1¶ Diagnostic IDs.
-
integer
id_t_predia= -1¶ Diagnostic IDs.
-
integer
id_s_predia= -1¶ Diagnostic IDs.
-
integer
id_e_predia= -1¶ Diagnostic IDs.
-
integer
id_diabatic_diff_temp_tend= -1¶ Diagnostic IDs.
-
integer
id_diabatic_diff_saln_tend= -1¶ Diagnostic IDs.
-
integer
id_diabatic_diff_heat_tend= -1¶ Diagnostic IDs.
-
integer
id_diabatic_diff_salt_tend= -1¶ Diagnostic IDs.
-
integer
id_diabatic_diff_heat_tend_2d= -1¶ Diagnostic IDs.
-
integer
id_diabatic_diff_salt_tend_2d= -1¶ Diagnostic IDs.
-
integer
id_diabatic_diff_h= -1¶ Diagnostic IDs.
-
integer
id_boundary_forcing_h= -1¶ Diagnostic IDs.
-
integer
id_boundary_forcing_h_tendency= -1¶ Diagnostic IDs.
-
integer
id_boundary_forcing_temp_tend= -1¶ Diagnostic IDs.
-
integer
id_boundary_forcing_saln_tend= -1¶ Diagnostic IDs.
-
integer
id_boundary_forcing_heat_tend= -1¶ Diagnostic IDs.
-
integer
id_boundary_forcing_salt_tend= -1¶ Diagnostic IDs.
-
integer
id_boundary_forcing_heat_tend_2d= -1¶ Diagnostic IDs.
-
integer
id_boundary_forcing_salt_tend_2d= -1¶ Diagnostic IDs.
-
integer
id_frazil_h= -1¶ Diagnostic IDs.
-
integer
id_frazil_temp_tend= -1¶ Diagnostic IDs.
-
integer
id_frazil_heat_tend= -1¶ Diagnostic IDs.
-
integer
id_frazil_heat_tend_2d= -1¶ Diagnostic IDs.
-
logical
diabatic_diff_tendency_diag= .false.¶ If true calculate diffusive tendency diagnostics.
-
logical
boundary_forcing_tendency_diag= .false.¶ If true calculate frazil diagnostics.
-
logical
frazil_tendency_diag= .false.¶ If true calculate frazil tendency diagnostics.
-
real, dimension(:,:,:), allocatable mom_diabatic_driver::diabatic_cs::frazil_heat_diag diagnose 3d heat tendency from frazil
-
real, dimension(:,:,:), allocatable mom_diabatic_driver::diabatic_cs::frazil_temp_diag diagnose 3d temp tendency from frazil
-
type(diabatic_aux_cs), pointer mom_diabatic_driver::diabatic_cs::diabatic_aux_csp => NULL() Control structure for a child module.
-
type(entrain_diffusive_cs), pointer mom_diabatic_driver::diabatic_cs::entrain_diffusive_csp => NULL() Control structure for a child module.
-
type(bulkmixedlayer_cs), pointer mom_diabatic_driver::diabatic_cs::bulkmixedlayer_csp => NULL() Control structure for a child module.
-
type(energetic_pbl_cs), pointer mom_diabatic_driver::diabatic_cs::energetic_pbl_csp => NULL() Control structure for a child module.
-
type(regularize_layers_cs), pointer mom_diabatic_driver::diabatic_cs::regularize_layers_csp => NULL() Control structure for a child module.
-
type(geothermal_cs), pointer mom_diabatic_driver::diabatic_cs::geothermal_csp => NULL() Control structure for a child module.
-
type(int_tide_cs), pointer mom_diabatic_driver::diabatic_cs::int_tide_csp => NULL() Control structure for a child module.
-
type(int_tide_input_cs), pointer mom_diabatic_driver::diabatic_cs::int_tide_input_csp => NULL() Control structure for a child module.
-
type(int_tide_input_type), pointer mom_diabatic_driver::diabatic_cs::int_tide_input => NULL() Control structure for a child module.
-
type(opacity_cs), pointer mom_diabatic_driver::diabatic_cs::opacity_csp => NULL() Control structure for a child module.
-
type(set_diffusivity_cs), pointer mom_diabatic_driver::diabatic_cs::set_diff_csp => NULL() Control structure for a child module.
-
type(sponge_cs), pointer mom_diabatic_driver::diabatic_cs::sponge_csp => NULL() Control structure for a child module.
-
type(ale_sponge_cs), pointer mom_diabatic_driver::diabatic_cs::ale_sponge_csp => NULL() Control structure for a child module.
-
type(tracer_flow_control_cs), pointer mom_diabatic_driver::diabatic_cs::tracer_flow_csp => NULL() Control structure for a child module.
-
type(optics_type), pointer mom_diabatic_driver::diabatic_cs::optics => NULL() Control structure for a child module.
-
type(kpp_cs), pointer mom_diabatic_driver::diabatic_cs::kpp_csp => NULL() Control structure for a child module.
-
type(tidal_mixing_cs), pointer mom_diabatic_driver::diabatic_cs::tidal_mixing_csp => NULL() Control structure for a child module.
-
type(cvmix_conv_cs), pointer mom_diabatic_driver::diabatic_cs::cvmix_conv_csp => NULL() Control structure for a child module.
-
type(diapyc_energy_req_cs), pointer mom_diabatic_driver::diabatic_cs::diapyc_en_rec_csp => NULL() Control structure for a child module.
-
type(group_pass_type) mom_diabatic_driver::diabatic_cs::pass_hold_eb_ea For group halo pass.
-
type(group_pass_type) mom_diabatic_driver::diabatic_cs::pass_kv For group halo pass.
-
type(diag_grid_storage) mom_diabatic_driver::diabatic_cs::diag_grids_prev Stores diagnostic grids at some previous point in the algorithm.
-
real, dimension(:,:,:), allocatable mom_diabatic_driver::diabatic_cs::kpp_nltheat KPP non-local transport for heat [m s-1].
-
real, dimension(:,:,:), allocatable mom_diabatic_driver::diabatic_cs::kpp_nltscalar KPP non-local transport for scalars [m s-1].
-
real, dimension(:,:,:), allocatable mom_diabatic_driver::diabatic_cs::kpp_buoy_flux KPP forcing buoyancy flux [L2 T-3 ~> m2 s-3].
-
real, dimension(:,:), allocatable mom_diabatic_driver::diabatic_cs::kpp_temp_flux KPP effective temperature flux [degC m s-1].
-
real, dimension(:,:), allocatable mom_diabatic_driver::diabatic_cs::kpp_salt_flux KPP effective salt flux [ppt m s-1].
-
type(time_type), pointer mom_diabatic_driver::diabatic_cs::time Pointer to model time (needed for sponges)
Public Members
-
logical
use_legacy_diabatic¶ If true (default), use the a legacy version of the diabatic algorithm. This is temporary and is needed to avoid change in answers.
-
logical
bulkmixedlayer¶ If true, a refined bulk mixed layer is used with nkml sublayers (and additional buffer layers).
-
logical
use_energetic_pbl¶ If true, use the implicit energetics planetary boundary layer scheme to determine the diffusivity in the surface boundary layer.
-
logical
use_kpp¶ If true, use CVMix/KPP boundary layer scheme to determine the OBLD and the diffusivities within this layer.
-
logical
use_kappa_shear¶ If true, use the kappa_shear module to find the shear-driven diapycnal diffusivity.
-
logical
use_cvmix_shear¶ If true, use the CVMix module to find the shear-driven diapycnal diffusivity.
-
logical
use_cvmix_ddiff¶ If true, use the CVMix double diffusion module.
-
logical
use_tidal_mixing¶ If true, activate tidal mixing diffusivity.
-
logical
use_cvmix_conv¶ If true, use the CVMix module to get enhanced mixing due to convection.
-
logical
use_sponge¶ If true, sponges may be applied anywhere in the domain. The exact location and properties of those sponges are set by calls to initialize_sponge and set_up_sponge_field.
-
logical
use_geothermal¶ If true, apply geothermal heating.
-
logical
use_int_tides¶ If true, use the code that advances a separate set of equations for the internal tide energy density.
-
logical
epbl_is_additive¶ If true, the diffusivity from ePBL is added to all other diffusivities. Otherwise, the larger of kappa- shear and ePBL diffusivities are used.
-
integer
nmode= 1¶ Number of baroclinic modes to consider.
-
real
uniform_test_cg¶ Uniform group velocity of internal tide for testing internal tides [m s-1] (BDM)
-
logical
usealealgorithm¶ If true, use the ALE algorithm rather than layered isopycnal/stacked shallow water mode. This logical passed by argument to diabatic_driver_init.
-
logical
aggregate_fw_forcing¶ Determines whether net incoming/outgoing surface FW fluxes are applied separately or combined before being applied.
-
real
ml_mix_first¶ The nondimensional fraction of the mixed layer algorithm that is applied before diffusive mixing. The default is 0, while 0.5 gives Strang splitting and 1 is a sensible value too. Note that if there are convective instabilities in the initial state, the first call may do much more than the second.
-
integer
nkbl¶ The number of buffer layers (if bulk_mixed_layer)
-
logical
massless_match_targets¶ If true (the default), keep the T & S consistent with the target values.
-
logical
mix_boundary_tracers¶ If true, mix the passive tracers in massless layers at the bottom into the interior as though a diffusivity of Kd_min_tr (see below) were operating.
-
real
kd_bbl_tr¶ A bottom boundary layer tracer diffusivity that will allow for explicitly specified bottom fluxes [Z2 T-1 ~> m2 s-1]. The entrainment at the bottom is at least sqrt(Kd_BBL_tr*dt) over the same distance.
-
real
kd_min_tr¶ A minimal diffusivity that should always be applied to tracers, especially in massless layers near the bottom [Z2 T-1 ~> m2 s-1].
-
real
minimum_forcing_depth= 0.001¶ The smallest depth over which heat and freshwater fluxes are applied [m].
-
real
evap_cfl_limit= 0.8¶ The largest fraction of a layer that can be evaporated in one time-step [nondim].
-
integer
halo_ts_diff= 0¶ The temperature, salinity and thickness halo size that must be valid for the diffusivity calculations.
-
logical
usekpp= .false.¶ use CVMix/KPP diffusivities and non-local transport
-
logical
salt_reject_below_ml¶ If true, add salt below mixed layer (layer mode only)
-
logical
kppispassive¶ If true, KPP is in passive mode, not changing answers.
-
logical
debug¶ If true, write verbose checksums for debugging purposes.
-
logical
debugconservation¶ If true, monitor conservation and extrema.
-
logical
tracer_tridiag¶ If true, use tracer_vertdiff instead of tridiagTS for vertical diffusion of T and S.
-
logical
debug_energy_req¶ If true, test the mixing energy requirement code.
-
type(diag_ctrl), pointer mom_diabatic_driver::diabatic_cs::diag structure used to regulate timing of diagnostic output
-
real
mlddensitydifference¶ Density difference used to determine MLD_user.
-
real
dz_subml_n2¶ The distance over which to calculate a diagnostic of the average stratification at the base of the mixed layer [Z ~> m].
-
integer
- type
The following data type a list of diagnostic fields an their variants, as well as variables that control the handling of model output.
Unnamed Group
-
integer
available_diag_doc_unit= -1¶ The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
integer
chksum_iounit= -1¶ The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
logical
diag_as_chksum¶ If true, log chksums in a text file instead of posting diagnostics.
-
integer
is¶ The start i-index of cell centers within the computational domain.
-
integer
ie¶ The end i-index of cell centers within the computational domain.
-
integer
js¶ The start j-index of cell centers within the computational domain.
-
integer
je¶ The end j-index of cell centers within the computational domain.
-
integer
isd¶ The start i-index of cell centers within the data domain.
-
integer
ied¶ The end i-index of cell centers within the data domain.
-
integer
jsd¶ The start j-index of cell centers within the data domain.
-
integer
jed¶ The end j-index of cell centers within the data domain.
-
real
time_int¶ The time interval for any fields that are offered for averaging [s].
-
type(time_type) mom_diag_mediator::diag_ctrl::time_end The end time of the valid interval for any offered field.
-
logical
ave_enabled= .false.¶ True if averaging is enabled.
-
type(axes_grp) mom_diag_mediator::diag_ctrl::axesbl The following are 3D and 2D axis groups defined for output. The names indicate the horizontal (B, T, Cu, or Cv) and vertical (L, i, or 1) locations.
-
type(axes_grp) mom_diag_mediator::diag_ctrl::axestl The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
type(axes_grp) mom_diag_mediator::diag_ctrl::axescul The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
type(axes_grp) mom_diag_mediator::diag_ctrl::axescvl The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
type(axes_grp) mom_diag_mediator::diag_ctrl::axesbi The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
type(axes_grp) mom_diag_mediator::diag_ctrl::axesti The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
type(axes_grp) mom_diag_mediator::diag_ctrl::axescui The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
type(axes_grp) mom_diag_mediator::diag_ctrl::axescvi The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
type(axes_grp) mom_diag_mediator::diag_ctrl::axesb1 The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
type(axes_grp) mom_diag_mediator::diag_ctrl::axest1 The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
type(axes_grp) mom_diag_mediator::diag_ctrl::axescu1 The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
type(axes_grp) mom_diag_mediator::diag_ctrl::axescv1 The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
type(axes_grp) mom_diag_mediator::diag_ctrl::axeszi A 1-D z-space axis at interfaces.
-
type(axes_grp) mom_diag_mediator::diag_ctrl::axeszl A 1-D z-space axis at layer centers.
-
type(axes_grp) mom_diag_mediator::diag_ctrl::axesnull An axis group for scalars.
-
real, dimension(:,:), pointer mom_diag_mediator::diag_ctrl::mask2dt => null() 2D mask array for cell-center points
-
real, dimension(:,:), pointer mom_diag_mediator::diag_ctrl::mask2dbu => null() 2D mask array for cell-corner points
-
real, dimension(:,:), pointer mom_diag_mediator::diag_ctrl::mask2dcu => null() 2D mask array for east-face points
-
real, dimension(:,:), pointer mom_diag_mediator::diag_ctrl::mask2dcv => null() 2D mask array for north-face points
-
real, dimension(:,:,:), pointer mom_diag_mediator::diag_ctrl::mask3dtl => null() 3D mask arrays for diagnostics at layers (mask…L) and interfaces (mask…i)
-
real, dimension(:,:,:), pointer mom_diag_mediator::diag_ctrl::mask3dbl => null() The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
real, dimension(:,:,:), pointer mom_diag_mediator::diag_ctrl::mask3dcul => null() The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
real, dimension(:,:,:), pointer mom_diag_mediator::diag_ctrl::mask3dcvl => null() The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
real, dimension(:,:,:), pointer mom_diag_mediator::diag_ctrl::mask3dti => null() The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
real, dimension(:,:,:), pointer mom_diag_mediator::diag_ctrl::mask3dbi => null() The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
real, dimension(:,:,:), pointer mom_diag_mediator::diag_ctrl::mask3dcui => null() The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
real, dimension(:,:,:), pointer mom_diag_mediator::diag_ctrl::mask3dcvi => null() The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
type(diagcs_dsamp), dimension(2:max_dsamp_lev) mom_diag_mediator::diag_ctrl::dsamp Downsample control container.
-
type(diag_type), dimension(:), allocatable mom_diag_mediator::diag_ctrl::diags The list of diagnostics.
-
integer
next_free_diag_id¶ The next unused diagnostic ID.
-
real
missing_value= -1.0e+34¶ default missing value to be sent to ALL diagnostics registrations
-
integer
num_diag_coords¶ Number of diagnostic vertical coordinates (remapped)
-
type(diag_remap_ctrl), dimension(:), allocatable mom_diag_mediator::diag_ctrl::diag_remap_cs Control structure for each possible coordinate.
-
type(diag_grid_storage) mom_diag_mediator::diag_ctrl::diag_grid_temp Stores the remapped diagnostic grid.
-
logical
diag_grid_overridden= .false.¶ True if the diagnostic grids have been overriden.
-
type(axes_grp), dimension(:), allocatable mom_diag_mediator::diag_ctrl::remap_axeszl The 1-D z-space cell-centered axis for remapping.
-
type(axes_grp), dimension(:), allocatable mom_diag_mediator::diag_ctrl::remap_axeszi The 1-D z-space interface axis for remapping.
-
type(axes_grp), dimension(:), allocatable mom_diag_mediator::diag_ctrl::remap_axestl The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
type(axes_grp), dimension(:), allocatable mom_diag_mediator::diag_ctrl::remap_axesbl The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
type(axes_grp), dimension(:), allocatable mom_diag_mediator::diag_ctrl::remap_axescul The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
type(axes_grp), dimension(:), allocatable mom_diag_mediator::diag_ctrl::remap_axescvl The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
type(axes_grp), dimension(:), allocatable mom_diag_mediator::diag_ctrl::remap_axesti The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
type(axes_grp), dimension(:), allocatable mom_diag_mediator::diag_ctrl::remap_axesbi The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
type(axes_grp), dimension(:), allocatable mom_diag_mediator::diag_ctrl::remap_axescui The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
type(axes_grp), dimension(:), allocatable mom_diag_mediator::diag_ctrl::remap_axescvi The unit number of a diagnostic documentation file. This file is open if available_diag_doc_unit is > 0.
-
real, dimension(:,:,:), pointer mom_diag_mediator::diag_ctrl::h => null() The thicknesses needed for remapping.
-
real, dimension(:,:,:), pointer mom_diag_mediator::diag_ctrl::t => null() The temperatures needed for remapping.
-
real, dimension(:,:,:), pointer mom_diag_mediator::diag_ctrl::s => null() The salinities needed for remapping.
-
type(eos_type), pointer mom_diag_mediator::diag_ctrl::eqn_of_state => null() The equation of state type.
-
type(ocean_grid_type), pointer mom_diag_mediator::diag_ctrl::g => null() The ocean grid type.
-
type(verticalgrid_type), pointer mom_diag_mediator::diag_ctrl::gv => null() The model’s vertical ocean grid.
-
type(unit_scale_type), pointer mom_diag_mediator::diag_ctrl::us => null() A dimensional unit scaling type.
-
integer
volume_cell_measure_dm_id= -1¶ The volume cell measure (special diagnostic) manager id.
-
integer
num_chksum_diags¶ Number of checksum-only diagnostics.
-
integer
- type
Contained for down sampled masks.
Private Members
-
real, dimension(:,:), pointer mom_diag_mediator::diag_dsamp::mask2d => null() Mask for 2d (x-y) axes.
-
real, dimension(:,:,:), pointer mom_diag_mediator::diag_dsamp::mask3d => null() Mask for 3d axes.
-
- type
Stores all the remapping grids and the model’s native space thicknesses.
Public Members
-
integer
num_diag_coords¶ Number of target coordinates.
-
real, dimension(:,:,:), allocatable mom_diag_mediator::diag_grid_storage::h_state Layer thicknesses in native space.
-
type(diag_grids_type), dimension(:), allocatable mom_diag_mediator::diag_grid_storage::diag_grids Primarily empty, except h field.
-
integer
- type
Contains an array to store a diagnostic target grid.
Private Members
-
real, dimension(:,:,:), allocatable mom_diag_mediator::diag_grids_type::h Target grid for remapped coordinate.
-
- type
Represents remapping of diagnostics to a particular vertical coordinate.
There is one of these types for each vertical coordinate. The vertical axes of a diagnostic will reference an instance of this type indicating how (or if) the diagnostic should be vertically remapped when being posted.
Private Members
-
logical
configured= .false.¶ Whether vertical coordinate has been configured.
-
logical
initialized= .false.¶ Whether remappping initialized.
-
logical
used= .false.¶ Whether this coordinate actually gets used.
-
integer
vertical_coord= 0¶ The vertical coordinate that we remap to.
-
character(len=10) mom_diag_remap::diag_remap_ctrl::vertical_coord_name ='' The coordinate name as understood by ALE.
-
character(len=16) mom_diag_remap::diag_remap_ctrl::diag_coord_name = '' A name for the purpose of run-time parameters.
-
character(len=8) mom_diag_remap::diag_remap_ctrl::diag_module_suffix = '' The suffix for the module to appear in diag_table.
-
type(remapping_cs) mom_diag_remap::diag_remap_ctrl::remap_cs Remapping control structure use for this axes.
-
type(regridding_cs) mom_diag_remap::diag_remap_ctrl::regrid_cs Regridding control structure that defines the coordiantes for this axes.
-
integer
nz= 0¶ Number of vertical levels used for remapping.
-
real, dimension(:,:,:), allocatable mom_diag_remap::diag_remap_ctrl::h Remap grid thicknesses.
-
real, dimension(:), allocatable mom_diag_remap::diag_remap_ctrl::dz Nominal layer thicknesses.
-
integer
interface_axes_id= 0¶ Vertical axes id for remapping at interfaces.
-
integer
layer_axes_id= 0¶ Vertical axes id for remapping on layers.
-
logical
- type
This type is used to represent a diagnostic at the diag_mediator level.
There can be both ‘primary’ and ‘seconday’ diagnostics. The primaries reside in the diag_csdiags array. They have an id which is an index into this array. The secondaries are ‘variations’ on the primary diagnostic. For example the CMOR diagnostics are secondary. The secondary diagnostics are kept in a list with the primary diagnostic as the head.
Private Members
-
logical
in_use¶ True if this entry is being used.
-
integer
fms_diag_id¶ Underlying FMS diag_manager id.
-
integer
fms_xyave_diag_id= -1¶ For a horizontally area-averaged diagnostic.
-
integer
downsample_diag_id= -1¶ For a horizontally area-downsampled diagnostic.
-
character(64) mom_diag_mediator::diag_type::debug_str = '' For FATAL errors and debugging.
-
type(axes_grp), pointer mom_diag_mediator::diag_type::axes => null() The axis group for this diagnostic.
-
type(diag_type), pointer mom_diag_mediator::diag_type::next => null() Pointer to the next diagnostic.
-
real
conversion_factor= 0.¶ A factor to multiply data by before posting to FMS, if non-zero.
-
logical
v_extensive= .false.¶ True for vertically extensive fields (vertically integrated). False for intensive (concentrations).
-
integer
xyz_method= 0¶ A 3 digit integer encoding the diagnostics cell method It can be used to determine the downsample algorithm.
-
logical
- type
Container for down sampling information.
Unnamed Group
-
type(axes_grp) mom_diag_mediator::diagcs_dsamp::axesbl Axes for each location on a diagnostic grid.
-
type(axes_grp) mom_diag_mediator::diagcs_dsamp::axestl Axes for each location on a diagnostic grid.
-
type(axes_grp) mom_diag_mediator::diagcs_dsamp::axescul Axes for each location on a diagnostic grid.
-
type(axes_grp) mom_diag_mediator::diagcs_dsamp::axescvl Axes for each location on a diagnostic grid.
-
type(axes_grp) mom_diag_mediator::diagcs_dsamp::axesbi Axes for each location on a diagnostic grid.
-
type(axes_grp) mom_diag_mediator::diagcs_dsamp::axesti Axes for each location on a diagnostic grid.
-
type(axes_grp) mom_diag_mediator::diagcs_dsamp::axescui Axes for each location on a diagnostic grid.
-
type(axes_grp) mom_diag_mediator::diagcs_dsamp::axescvi Axes for each location on a diagnostic grid.
-
type(axes_grp) mom_diag_mediator::diagcs_dsamp::axesb1 Axes for each location on a diagnostic grid.
-
type(axes_grp) mom_diag_mediator::diagcs_dsamp::axest1 Axes for each location on a diagnostic grid.
-
type(axes_grp) mom_diag_mediator::diagcs_dsamp::axescu1 Axes for each location on a diagnostic grid.
-
type(axes_grp) mom_diag_mediator::diagcs_dsamp::axescv1 Axes for each location on a diagnostic grid.
-
type(axes_grp), dimension(:), allocatable mom_diag_mediator::diagcs_dsamp::remap_axestl Axes for each location on a diagnostic grid.
-
type(axes_grp), dimension(:), allocatable mom_diag_mediator::diagcs_dsamp::remap_axesbl Axes for each location on a diagnostic grid.
-
type(axes_grp), dimension(:), allocatable mom_diag_mediator::diagcs_dsamp::remap_axescul Axes for each location on a diagnostic grid.
-
type(axes_grp), dimension(:), allocatable mom_diag_mediator::diagcs_dsamp::remap_axescvl Axes for each location on a diagnostic grid.
-
type(axes_grp), dimension(:), allocatable mom_diag_mediator::diagcs_dsamp::remap_axesti Axes for each location on a diagnostic grid.
-
type(axes_grp), dimension(:), allocatable mom_diag_mediator::diagcs_dsamp::remap_axesbi Axes for each location on a diagnostic grid.
-
type(axes_grp), dimension(:), allocatable mom_diag_mediator::diagcs_dsamp::remap_axescui Axes for each location on a diagnostic grid.
-
type(axes_grp), dimension(:), allocatable mom_diag_mediator::diagcs_dsamp::remap_axescvi Axes for each location on a diagnostic grid.
-
real, dimension(:,:), pointer mom_diag_mediator::diagcs_dsamp::mask2dt => null() 2D mask array for cell-center points
-
real, dimension(:,:), pointer mom_diag_mediator::diagcs_dsamp::mask2dbu => null() 2D mask array for cell-corner points
-
real, dimension(:,:), pointer mom_diag_mediator::diagcs_dsamp::mask2dcu => null() 2D mask array for east-face points
-
real, dimension(:,:), pointer mom_diag_mediator::diagcs_dsamp::mask2dcv => null() 2D mask array for north-face points
-
real, dimension(:,:,:), pointer mom_diag_mediator::diagcs_dsamp::mask3dtl => null() 3D mask arrays for diagnostics at layers (mask…L) and interfaces (mask…i)
-
real, dimension(:,:,:), pointer mom_diag_mediator::diagcs_dsamp::mask3dbl => null() Axes for each location on a diagnostic grid.
-
real, dimension(:,:,:), pointer mom_diag_mediator::diagcs_dsamp::mask3dcul => null() Axes for each location on a diagnostic grid.
-
real, dimension(:,:,:), pointer mom_diag_mediator::diagcs_dsamp::mask3dcvl => null() Axes for each location on a diagnostic grid.
-
real, dimension(:,:,:), pointer mom_diag_mediator::diagcs_dsamp::mask3dti => null() Axes for each location on a diagnostic grid.
-
real, dimension(:,:,:), pointer mom_diag_mediator::diagcs_dsamp::mask3dbi => null() Axes for each location on a diagnostic grid.
-
real, dimension(:,:,:), pointer mom_diag_mediator::diagcs_dsamp::mask3dcui => null() Axes for each location on a diagnostic grid.
-
real, dimension(:,:,:), pointer mom_diag_mediator::diagcs_dsamp::mask3dcvi => null() Axes for each location on a diagnostic grid.
Private Members
-
integer
isc¶ The start i-index of cell centers within the computational domain.
-
integer
iec¶ The end i-index of cell centers within the computational domain.
-
integer
jsc¶ The start j-index of cell centers within the computational domain.
-
integer
jec¶ The end j-index of cell centers within the computational domain.
-
integer
isd¶ The start i-index of cell centers within the data domain.
-
integer
ied¶ The end i-index of cell centers within the data domain.
-
integer
jsd¶ The start j-index of cell centers within the data domain.
-
integer
jed¶ The end j-index of cell centers within the data domain.
-
integer
isg¶ The start i-index of cell centers within the global domain.
-
integer
ieg¶ The end i-index of cell centers within the global domain.
-
integer
jsg¶ The start j-index of cell centers within the global domain.
-
integer
jeg¶ The end j-index of cell centers within the global domain.
-
integer
isgb¶ The start i-index of cell corners within the global domain.
-
integer
iegb¶ The end i-index of cell corners within the global domain.
-
integer
jsgb¶ The start j-index of cell corners within the global domain.
-
integer
jegb¶ The end j-index of cell corners within the global domain.
-
- type
The control structure for the MOM_diagnostics module.
Unnamed Group
-
integer
id_u= -1¶ Diagnostic IDs.
-
integer
id_v= -1¶ Diagnostic IDs.
-
integer
id_h= -1¶ Diagnostic IDs.
-
integer
id_e= -1¶ Diagnostic IDs.
-
integer
id_e_d= -1¶ Diagnostic IDs.
-
integer
id_du_dt= -1¶ Diagnostic IDs.
-
integer
id_dv_dt= -1¶ Diagnostic IDs.
-
integer
id_col_ht= -1¶ Diagnostic IDs.
-
integer
id_dh_dt= -1¶ Diagnostic IDs.
-
integer
id_ke= -1¶ Diagnostic IDs.
-
integer
id_dkedt= -1¶ Diagnostic IDs.
-
integer
id_pe_to_ke= -1¶ Diagnostic IDs.
-
integer
id_ke_coradv= -1¶ Diagnostic IDs.
-
integer
id_ke_adv= -1¶ Diagnostic IDs.
-
integer
id_ke_visc= -1¶ Diagnostic IDs.
-
integer
id_ke_horvisc= -1¶ Diagnostic IDs.
-
integer
id_ke_dia= -1¶ Diagnostic IDs.
-
integer
id_uh_rlay= -1¶ Diagnostic IDs.
-
integer
id_vh_rlay= -1¶ Diagnostic IDs.
-
integer
id_uhgm_rlay= -1¶ Diagnostic IDs.
-
integer
id_vhgm_rlay= -1¶ Diagnostic IDs.
-
integer
id_h_rlay= -1¶ Diagnostic IDs.
-
integer
id_rd1= -1¶ Diagnostic IDs.
-
integer
id_rml= -1¶ Diagnostic IDs.
-
integer
id_rcv= -1¶ Diagnostic IDs.
-
integer
id_cg1= -1¶ Diagnostic IDs.
-
integer
id_cfl_cg1= -1¶ Diagnostic IDs.
-
integer
id_cfl_cg1_x= -1¶ Diagnostic IDs.
-
integer
id_cfl_cg1_y= -1¶ Diagnostic IDs.
-
integer
id_cg_ebt= -1¶ Diagnostic IDs.
-
integer
id_rd_ebt= -1¶ Diagnostic IDs.
-
integer
id_p_ebt= -1¶ Diagnostic IDs.
-
integer
id_temp_int= -1¶ Diagnostic IDs.
-
integer
id_salt_int= -1¶ Diagnostic IDs.
-
integer
id_mass_wt= -1¶ Diagnostic IDs.
-
integer
id_col_mass= -1¶ Diagnostic IDs.
-
integer
id_masscello= -1¶ Diagnostic IDs.
-
integer
id_masso= -1¶ Diagnostic IDs.
-
integer
id_volcello= -1¶ Diagnostic IDs.
-
integer
id_tpot= -1¶ Diagnostic IDs.
-
integer
id_sprac= -1¶ Diagnostic IDs.
-
integer
id_tob= -1¶ Diagnostic IDs.
-
integer
id_sob= -1¶ Diagnostic IDs.
-
integer
id_thetaoga= -1¶ Diagnostic IDs.
-
integer
id_soga= -1¶ Diagnostic IDs.
-
integer
id_sosga= -1¶ Diagnostic IDs.
-
integer
id_tosga= -1¶ Diagnostic IDs.
-
integer
id_temp_layer_ave= -1¶ Diagnostic IDs.
-
integer
id_salt_layer_ave= -1¶ Diagnostic IDs.
-
integer
id_pbo= -1¶ Diagnostic IDs.
-
integer
id_thkcello= -1¶ Diagnostic IDs.
-
integer
id_rhoinsitu= -1¶ Diagnostic IDs.
-
integer
id_rhopot0= -1¶ Diagnostic IDs.
-
integer
id_rhopot2= -1¶ Diagnostic IDs.
-
integer
id_h_pre_sync= -1¶ Diagnostic IDs.
-
type(wave_speed_cs), pointer mom_diagnostics::diagnostics_cs::wave_speed_csp => NULL() The control structure for calculating wave speed.
-
type(p3d), dimension( 50 ) mom_diagnostics::diagnostics_cs::var_ptr pointers to variables used in the calculation of time derivatives
-
type(p3d), dimension( 50 ) mom_diagnostics::diagnostics_cs::deriv Time derivatives of various fields.
-
type(p3d), dimension( 50 ) mom_diagnostics::diagnostics_cs::prev_val Previous values of variables used in the calculation of time derivatives previous values of variables used in calculation of time derivatives.
-
integer, dimension( 50 ) mom_diagnostics::diagnostics_cs::nlay The number of layers in each diagnostics.
-
integer
num_time_deriv= 0¶ The number of time derivative diagnostics.
-
type(group_pass_type) mom_diagnostics::diagnostics_cs::pass_ke_uv A handle used for group halo passes.
Public Members
-
real
mono_n2_column_fraction= 0.¶ The lower fraction of water column over which N2 is limited as monotonic for the purposes of calculating the equivalent barotropic wave speed.
-
real
mono_n2_depth= -1.¶ The depth below which N2 is limited as monotonic for the purposes of calculating the equivalent barotropic wave speed [m].
-
type(diag_ctrl), pointer mom_diagnostics::diagnostics_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
real, dimension(:,:,:), pointer mom_diagnostics::diagnostics_cs::e => NULL() interface height [Z ~> m]
-
real, dimension(:,:,:), pointer mom_diagnostics::diagnostics_cs::e_d => NULL() interface height above bottom [Z ~> m]
-
real, dimension(:,:,:), pointer mom_diagnostics::diagnostics_cs::du_dt => NULL() net i-acceleration [m s-2]
-
real, dimension(:,:,:), pointer mom_diagnostics::diagnostics_cs::dv_dt => NULL() net j-acceleration [m s-2]
-
real, dimension(:,:,:), pointer mom_diagnostics::diagnostics_cs::dh_dt => NULL() thickness rate of change [H s-1 ~> m s-1 or kg m-2 s-1]
-
real, dimension(:,:,:), pointer mom_diagnostics::diagnostics_cs::p_ebt => NULL() Equivalent barotropic modal structure [nondim].
-
real, dimension(:,:,:), pointer mom_diagnostics::diagnostics_cs::h_rlay => NULL() Layer thicknesses in potential density coordinates [H ~> m or kg m-2].
-
real, dimension(:,:,:), pointer mom_diagnostics::diagnostics_cs::uh_rlay => NULL() Zonal transports in potential density coordinates [H m2 s-1 ~> m3 s-1 or kg s-1].
-
real, dimension(:,:,:), pointer mom_diagnostics::diagnostics_cs::vh_rlay => NULL() Meridional transports in potential density coordinates [H m2 s-1 ~> m3 s-1 or kg s-1].
-
real, dimension(:,:,:), pointer mom_diagnostics::diagnostics_cs::uhgm_rlay => NULL() Zonal Gent-McWilliams transports in potential density coordinates [H m2 s-1 ~> m3 s-1 or kg s-1].
-
real, dimension(:,:,:), pointer mom_diagnostics::diagnostics_cs::vhgm_rlay => NULL() Meridional Gent-McWilliams transports in potential density coordinates [H m2 s-1 ~> m3 s-1 or kg s-1].
-
real, dimension(:,:), pointer mom_diagnostics::diagnostics_cs::cg1 => NULL() First baroclinic gravity wave speed [m s-1].
-
real, dimension(:,:), pointer mom_diagnostics::diagnostics_cs::rd1 => NULL() First baroclinic deformation radius [m].
-
real, dimension(:,:), pointer mom_diagnostics::diagnostics_cs::cfl_cg1 => NULL() CFL for first baroclinic gravity wave speed, nondim.
-
real, dimension(:,:), pointer mom_diagnostics::diagnostics_cs::cfl_cg1_x => NULL() i-component of CFL for first baroclinic gravity wave speed, nondim
-
real, dimension(:,:), pointer mom_diagnostics::diagnostics_cs::cfl_cg1_y => NULL() j-component of CFL for first baroclinic gravity wave speed, nondim
-
real, dimension(:,:,:), pointer mom_diagnostics::diagnostics_cs::ke => NULL() KE per unit mass [m2 s-2].
-
real, dimension(:,:,:), pointer mom_diagnostics::diagnostics_cs::dke_dt => NULL() time derivative of the layer KE [m3 s-3]
-
real, dimension(:,:,:), pointer mom_diagnostics::diagnostics_cs::pe_to_ke => NULL() potential energy to KE term [m3 s-3]
-
real, dimension(:,:,:), pointer mom_diagnostics::diagnostics_cs::ke_coradv => NULL() KE source from the combined Coriolis and advection terms [m3 s-3].
-
real, dimension(:,:,:), pointer mom_diagnostics::diagnostics_cs::ke_adv => NULL() KE source from along-layer advection [m3 s-3].
-
real, dimension(:,:,:), pointer mom_diagnostics::diagnostics_cs::ke_visc => NULL() KE source from vertical viscosity [m3 s-3].
-
real, dimension(:,:,:), pointer mom_diagnostics::diagnostics_cs::ke_horvisc => NULL() KE source from horizontal viscosity [m3 s-3].
-
real, dimension(:,:,:), pointer mom_diagnostics::diagnostics_cs::ke_dia => NULL() KE source from diapycnal diffusion [m3 s-3].
-
integer
- type
This control structure holds parameters for the MOM_diapyc_energy_req module.
Unnamed Group
-
integer
id_ert= -1¶ Diagnostic IDs.
-
integer
id_erb= -1¶ Diagnostic IDs.
-
integer
id_erc= -1¶ Diagnostic IDs.
-
integer
id_erh= -1¶ Diagnostic IDs.
-
integer
id_kddt= -1¶ Diagnostic IDs.
-
integer
id_kd= -1¶ Diagnostic IDs.
-
integer
id_chct= -1¶ Diagnostic IDs.
-
integer
id_chcb= -1¶ Diagnostic IDs.
-
integer
id_chcc= -1¶ Diagnostic IDs.
-
integer
id_chch= -1¶ Diagnostic IDs.
-
integer
id_t0= -1¶ Diagnostic IDs.
-
integer
id_tf= -1¶ Diagnostic IDs.
-
integer
id_s0= -1¶ Diagnostic IDs.
-
integer
id_sf= -1¶ Diagnostic IDs.
-
integer
id_n2_0= -1¶ Diagnostic IDs.
-
integer
id_n2_f= -1¶ Diagnostic IDs.
-
integer
id_h= -1¶ Diagnostic IDs.
-
integer
id_zint= -1¶ Diagnostic IDs.
Public Members
-
logical
initialized= .false.¶ A variable that is here because empty structures are not permitted by some compilers.
-
real
test_kh_scaling¶ A scaling factor for the diapycnal diffusivity.
-
real
colht_scaling¶ A scaling factor for the column height change correction term.
-
logical
use_test_kh_profile¶ If true, use the internal test diffusivity profile in place of any that might be passed in as an argument.
-
type(diag_ctrl), pointer mom_diapyc_energy_req::diapyc_energy_req_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
integer
- type
This structure has memory for used in calculating diagnostics of diffusivity.
Unnamed Group
-
real, dimension(:,:,:), pointer mom_set_diffusivity::diffusivity_diags::n2_3d => NULL() squared buoyancy frequency at interfaces [T-2 ~> s-2]
-
real, dimension(:,:,:), pointer mom_set_diffusivity::diffusivity_diags::kd_user => NULL() user-added diffusivity at interfaces [Z2 T-1 ~> m2 s-1]
-
real, dimension(:,:,:), pointer mom_set_diffusivity::diffusivity_diags::kd_bbl => NULL() BBL diffusivity at interfaces [Z2 T-1 ~> m2 s-1].
-
real, dimension(:,:,:), pointer mom_set_diffusivity::diffusivity_diags::kd_work => NULL() layer integrated work by diapycnal mixing [kg Z3 m-3 T-3 ~> W m-2]
-
real, dimension(:,:,:), pointer mom_set_diffusivity::diffusivity_diags::maxtke => NULL() energy required to entrain to h_max [Z3 T-3 ~> m3 s-3]
-
real, dimension(:,:,:), pointer mom_set_diffusivity::diffusivity_diags::kt_extra => NULL() double diffusion diffusivity for temp [Z2 T-1 ~> m2 s-1].
-
real, dimension(:,:,:), pointer mom_set_diffusivity::diffusivity_diags::ks_extra => NULL() double diffusion diffusivity for saln [Z2 T-1 ~> m2 s-1].
-
real, dimension(:,:,:), pointer mom_set_diffusivity::diffusivity_diags::tke_to_kd => NULL() conversion rate (~1.0 / (G_Earth + dRho_lay)) between TKE dissipated within a layer and Kd in that layer [Z2 T-1 / Z3 T-3 = T2 Z-1 ~> s2 m-1]
-
- type
Container for paths and parameter file names.
Public Members
-
character(len=240) mom_get_input::directories::restart_input_dir = ' ' The directory to read restart and input files.
-
character(len=240) mom_get_input::directories::restart_output_dir = ' ' The directory into which to write restart files.
-
character(len=240) mom_get_input::directories::output_directory = ' ' The directory to use to write the model output.
-
character(len=240) mom_get_input::directories::input_filename = ' ' A string that indicates the input files or how.
-
-
interface
doc_param¶ Document parameter values.
Private Functions
-
subroutine
doc_param_none(doc doc, varname varname, desc desc, units units)¶ This subroutine handles parameter documentation with no value.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] varname: The name of the parameter being documented[in] desc: A description of the parameter being documented[in] units: The units of the parameter being documented
-
subroutine mom_document::doc_param::doc_param_logical(doc doc, varname varname, desc desc, units units, val val, default default, layoutParam layoutParam, debuggingParam debuggingParam) This subroutine handles parameter documentation for logicals.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] varname: The name of the parameter being documented[in] desc: A description of the parameter being documented[in] units: The units of the parameter being documented[in] val: The value of this parameter[in] default: The default value of this parameter[in] layoutparam: If present and true, this is a layout parameter.[in] debuggingparam: If present and true, this is a debugging parameter.
-
subroutine mom_document::doc_param::doc_param_logical_array(doc doc, varname varname, desc desc, units units, vals vals, default default, layoutParam layoutParam, debuggingParam debuggingParam) This subroutine handles parameter documentation for arrays of logicals.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] varname: The name of the parameter being documented[in] desc: A description of the parameter being documented[in] units: The units of the parameter being documented[in] vals: The array of values to record[in] default: The default value of this parameter[in] layoutparam: If present and true, this is a layout parameter.[in] debuggingparam: If present and true, this is a debugging parameter.
-
subroutine mom_document::doc_param::doc_param_int(doc doc, varname varname, desc desc, units units, val val, default default, layoutParam layoutParam, debuggingParam debuggingParam) This subroutine handles parameter documentation for integers.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] varname: The name of the parameter being documented[in] desc: A description of the parameter being documented[in] units: The units of the parameter being documented[in] val: The value of this parameter[in] default: The default value of this parameter[in] layoutparam: If present and true, this is a layout parameter.[in] debuggingparam: If present and true, this is a debugging parameter.
-
subroutine mom_document::doc_param::doc_param_int_array(doc doc, varname varname, desc desc, units units, vals vals, default default, layoutParam layoutParam, debuggingParam debuggingParam) This subroutine handles parameter documentation for arrays of integers.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] varname: The name of the parameter being documented[in] desc: A description of the parameter being documented[in] units: The units of the parameter being documented[in] vals: The array of values to record[in] default: The default value of this parameter[in] layoutparam: If present and true, this is a layout parameter.[in] debuggingparam: If present and true, this is a debugging parameter.
-
subroutine mom_document::doc_param::doc_param_real(doc doc, varname varname, desc desc, units units, val val, default default, debuggingParam debuggingParam) This subroutine handles parameter documentation for reals.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] varname: The name of the parameter being documented[in] desc: A description of the parameter being documented[in] units: The units of the parameter being documented[in] val: The value of this parameter[in] default: The default value of this parameter[in] debuggingparam: If present and true, this is a debugging parameter.
-
subroutine mom_document::doc_param::doc_param_real_array(doc doc, varname varname, desc desc, units units, vals vals, default default, debuggingParam debuggingParam) This subroutine handles parameter documentation for arrays of reals.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] varname: The name of the parameter being documented[in] desc: A description of the parameter being documented[in] units: The units of the parameter being documented[in] vals: The array of values to record[in] default: The default value of this parameter[in] debuggingparam: If present and true, this is a debugging parameter.
-
subroutine mom_document::doc_param::doc_param_char(doc doc, varname varname, desc desc, units units, val val, default default, layoutParam layoutParam, debuggingParam debuggingParam) This subroutine handles parameter documentation for character strings.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] varname: The name of the parameter being documented[in] desc: A description of the parameter being documented[in] units: The units of the parameter being documented[in] val: The value of the parameter[in] default: The default value of this parameter[in] layoutparam: If present and true, this is a layout parameter.[in] debuggingparam: If present and true, this is a debugging parameter.
-
subroutine mom_document::doc_param::doc_param_time(doc doc, varname varname, desc desc, units units, val val, default default, layoutParam layoutParam, debuggingParam debuggingParam) This subroutine handles parameter documentation for time-type variables.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] varname: The name of the parameter being documented[in] desc: A description of the parameter being documented[in] units: The units of the parameter being documented[in] val: The value of the parameter[in] default: The default value of this parameter[in] layoutparam: If present and true, this is a layout parameter.[in] debuggingparam: If present and true, this is a debugging parameter.
-
subroutine
- type
A structure that controls where the documentation occurs, its veborsity and formatting.
Public Members
-
integer
unitall= -1¶ The open unit number for docFileBase + .all.
-
integer
unitshort= -1¶ The open unit number for docFileBase + .short.
-
integer
unitlayout= -1¶ The open unit number for docFileBase + .layout.
-
integer
unitdebugging= -1¶ The open unit number for docFileBase + .debugging.
-
logical
filesareopen= .false.¶ True if any files were successfully opened.
-
character(len=mlen) mom_document::doc_type::docfilebase = '' The basename of the files where run-time parameters, settings and defaults are documented.
-
logical
complete= .true.¶ If true, document all parameters.
-
logical
minimal= .true.¶ If true, document non-default parameters.
-
logical
layout= .true.¶ If true, document layout parameters.
-
logical
debugging= .true.¶ If true, document debugging parameters.
-
logical
definesyntax= .false.¶ If true, use ‘#def’ syntax instead of a=b syntax.
-
logical
warnonconflicts= .false.¶ Cause a WARNING error if defaults differ.
-
integer
commentcolumn= 32¶ Number of spaces before the comment marker.
-
integer
max_line_len= 112¶ The maximum length of message lines.
-
type(link_msg), pointer mom_document::doc_type::chain_msg => NULL() Database of messages.
-
character(len=240) mom_document::doc_type::blockprefix = '' The full name of the current block.
-
integer
- type
The DOME_tracer control structure.
Public Members
-
logical
coupled_tracers= .false.¶ These tracers are not offered to the coupler.
-
character(len=200) dome_tracer::dome_tracer_cs::tracer_ic_file The full path to the IC file, or ” ” to initialize internally.
-
type(time_type), pointer dome_tracer::dome_tracer_cs::time => NULL() A pointer to the ocean model’s clock.
-
type(tracer_registry_type), pointer dome_tracer::dome_tracer_cs::tr_reg => NULL() A pointer to the tracer registry.
-
real, dimension(:,:,:,:), pointer dome_tracer::dome_tracer_cs::tr => NULL() The array of tracers used in this package, in g m-3?
-
real, dimension(ntr) dome_tracer::dome_tracer_cs::land_val = -1.0 The value of tr used where land is masked out.
-
logical
use_sponge¶ If true, sponges may be applied somewhere in the domain.
-
integer, dimension(ntr) dome_tracer::dome_tracer_cs::ind_tr Indices returned by aof_set_coupler_flux if it is used and the surface tracer concentrations are to be provided to the coupler.
-
type(diag_ctrl), pointer dome_tracer::dome_tracer_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
type(vardesc), dimension(ntr) dome_tracer::dome_tracer_cs::tr_desc Descriptions and metadata for the tracers.
-
logical
-
interface
downsample_diag_field¶ Down sample a diagnostic field.
Private Functions
-
subroutine
downsample_diag_field_2d(locfield locfield, locfield_dsamp locfield_dsamp, dl dl, diag_cs diag_cs, diag diag, isv isv, iev iev, jsv jsv, jev jev, mask mask)¶ This subroutine allocates and computes a downsampled array from an input array It also determines the diagnostics-compurte indices for the downsampled array 2d interface.
- Parameters
locfield: Input array pointer[inout] locfield_dsamp: Output (downsampled) array[in] diag_cs: Structure used to regulate diagnostic output[in] diag: A structure describing the diagnostic to post[in] dl: Level of down sampling[inout] isv: i-start index for diagnostics[inout] iev: i-end index for diagnostics[inout] jsv: j-start index for diagnostics[inout] jev: j-end index for diagnostics[in] mask: If present, use this real array as the data mask.
-
subroutine
downsample_diag_field_3d(locfield locfield, locfield_dsamp locfield_dsamp, dl dl, diag_cs diag_cs, diag diag, isv isv, iev iev, jsv jsv, jev jev, mask mask)¶ This subroutine allocates and computes a downsampled array from an input array It also determines the diagnostics-compurte indices for the downsampled array 3d interface.
- Parameters
locfield: Input array pointer[inout] locfield_dsamp: Output (downsampled) array[in] diag_cs: Structure used to regulate diagnostic output[in] diag: A structure describing the diagnostic to post[in] dl: Level of down sampling[inout] isv: i-start index for diagnostics[inout] iev: i-end index for diagnostics[inout] jsv: j-start index for diagnostics[inout] jev: j-end index for diagnostics[in] mask: If present, use this real array as the data mask.
-
subroutine
-
interface
downsample_field¶ Down sample a field.
Private Functions
-
subroutine
downsample_field_2d(field_in field_in, field_out field_out, dl dl, method method, mask mask, diag_cs diag_cs, diag diag, isv_o isv_o, jsv_o jsv_o, isv_d isv_d, iev_d iev_d, jsv_d jsv_d, jev_d jev_d)¶ This subroutine allocates and computes a down sampled 2d array given an input array The down sample method is based on the “cell_methods” for the diagnostics as explained in the above table.
- Parameters
field_in: Original field to be down sampledfield_out: Down sampled field[in] dl: Level of down sampling[in] method: Sampling methodmask: Mask for field[in] diag_cs: Structure used to regulate diagnostic output[in] diag: A structure describing the diagnostic to post[in] isv_o: Original i-start index[in] jsv_o: Original j-start index[in] isv_d: i-start index of down sampled data[in] iev_d: i-end index of down sampled data[in] jsv_d: j-start index of down sampled data[in] jev_d: j-end index of down sampled data
-
subroutine
downsample_field_3d(field_in field_in, field_out field_out, dl dl, method method, mask mask, diag_cs diag_cs, diag diag, isv_o isv_o, jsv_o jsv_o, isv_d isv_d, iev_d iev_d, jsv_d jsv_d, jev_d jev_d)¶ This subroutine allocates and computes a down sampled 3d array given an input array The down sample method is based on the “cell_methods” for the diagnostics as explained in the above table.
-
subroutine
-
interface
downsample_mask¶ Down sample the mask of a field.
Private Functions
-
subroutine
downsample_mask_2d(field_in field_in, field_out field_out, dl dl, isc_o isc_o, jsc_o jsc_o, isc_d isc_d, iec_d iec_d, jsc_d jsc_d, jec_d jec_d, isd_d isd_d, ied_d ied_d, jsd_d jsd_d, jed_d jed_d)¶ Allocate and compute the 2d down sampled mask The masks are down sampled based on a minority rule, i.e., a coarse cell is open (1) if at least one of the sub-cells are open, otherwise it’s closed (0)
- Parameters
[in] field_in: Original field to be down sampledfield_out: Down sampled field[in] dl: Level of down sampling[in] isc_o: Original i-start index[in] jsc_o: Original j-start index[in] isc_d: Computational i-start index of down sampled data[in] iec_d: Computational i-end index of down sampled data[in] jsc_d: Computational j-start index of down sampled data[in] jec_d: Computational j-end index of down sampled data[in] isd_d: Computational i-start index of down sampled data[in] ied_d: Computational i-end index of down sampled data[in] jsd_d: Computational j-start index of down sampled data[in] jed_d: Computational j-end index of down sampled data
-
subroutine
downsample_mask_3d(field_in field_in, field_out field_out, dl dl, isc_o isc_o, jsc_o jsc_o, isc_d isc_d, iec_d iec_d, jsc_d jsc_d, jec_d jec_d, isd_d isd_d, ied_d ied_d, jsd_d jsd_d, jed_d jed_d)¶ Allocate and compute the 3d down sampled mask The masks are down sampled based on a minority rule, i.e., a coarse cell is open (1) if at least one of the sub-cells are open, otherwise it’s closed (0)
- Parameters
[in] field_in: Original field to be down sampledfield_out: down sampled field[in] dl: Level of down sampling[in] isc_o: Original i-start index[in] jsc_o: Original j-start index[in] isc_d: Computational i-start index of down sampled data[in] iec_d: Computational i-end index of down sampled data[in] jsc_d: Computational j-start index of down sampled data[in] jec_d: Computational j-end index of down sampled data[in] isd_d: Computational i-start index of down sampled data[in] ied_d: Computational i-end index of down sampled data[in] jsd_d: Computational j-start index of down sampled data[in] jed_d: Computational j-end index of down sampled data
-
subroutine
- type
Control structure for the dumbbell test case forcing.
Public Members
-
logical
use_temperature¶ If true, temperature and salinity are used as state variables.
-
logical
restorebuoy¶ If true, use restoring surface buoyancy forcing.
-
real
rho0¶ The density used in the Boussinesq approximation [kg m-3].
-
real
g_earth¶ The gravitational acceleration [L2 Z-1 T-2 ~> m s-2].
-
real
flux_const¶ The restoring rate at the surface [m s-1].
-
real
gust_const¶ A constant unresolved background gustiness that contributes to ustar [Pa].
-
real
slp_amplitude¶ The amplitude of pressure loading [Pa] applied to the reservoirs.
-
real
slp_period¶ Period of sinusoidal pressure wave.
-
real, dimension(:,:), allocatable dumbbell_surface_forcing::dumbbell_surface_forcing_cs::forcing_mask A mask regulating where forcing occurs.
-
real, dimension(:,:), allocatable dumbbell_surface_forcing::dumbbell_surface_forcing_cs::s_restore The surface salinity field toward which to.
-
type(diag_ctrl), pointer dumbbell_surface_forcing::dumbbell_surface_forcing_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
logical
- type
The control structure for the regional dyes tracer package.
Public Members
-
integer
ntr¶ The number of tracers that are actually used.
-
logical
coupled_tracers= .false.¶ These tracers are not offered to the coupler.
-
real, dimension(:), allocatable regional_dyes::dye_tracer_cs::dye_source_minlon Minimum longitude of region dye will be injected.
-
real, dimension(:), allocatable regional_dyes::dye_tracer_cs::dye_source_maxlon Maximum longitude of region dye will be injected.
-
real, dimension(:), allocatable regional_dyes::dye_tracer_cs::dye_source_minlat Minimum latitude of region dye will be injected.
-
real, dimension(:), allocatable regional_dyes::dye_tracer_cs::dye_source_maxlat Maximum latitude of region dye will be injected.
-
real, dimension(:), allocatable regional_dyes::dye_tracer_cs::dye_source_mindepth Minimum depth of region dye will be injected [Z ~> m].
-
real, dimension(:), allocatable regional_dyes::dye_tracer_cs::dye_source_maxdepth Maximum depth of region dye will be injected [Z ~> m].
-
type(tracer_registry_type), pointer regional_dyes::dye_tracer_cs::tr_reg => NULL() A pointer to the tracer registry.
-
real, dimension(:,:,:,:), pointer regional_dyes::dye_tracer_cs::tr => NULL() The array of tracers used in this subroutine, in g m-3?
-
integer, dimension(:), allocatable regional_dyes::dye_tracer_cs::ind_tr Indices returned by aof_set_coupler_flux if it is used and the surface tracer concentrations are to be provided to the coupler.
-
type(diag_ctrl), pointer regional_dyes::dye_tracer_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
type(mom_restart_cs), pointer regional_dyes::dye_tracer_cs::restart_csp => NULL() A pointer to the restart control structure.
-
type(vardesc), dimension(:), allocatable regional_dyes::dye_tracer_cs::tr_desc Descriptions and metadata for the tracers.
-
logical
tracers_may_reinit= .false.¶ If true the tracers may be initialized if not found in a restart file.
-
integer
- type
Control structure for dyed-channel open boundaries.
- type
The control structure for the dyed_obc tracer package.
Public Members
-
integer
ntr¶ The number of tracers that are actually used.
-
logical
coupled_tracers= .false.¶ These tracers are not offered to the coupler.
-
character(len=200) dyed_obc_tracer::dyed_obc_tracer_cs::tracer_ic_file The full path to the IC file, or ” ” to initialize internally.
-
type(time_type), pointer dyed_obc_tracer::dyed_obc_tracer_cs::time => NULL() A pointer to the ocean model’s clock.
-
type(tracer_registry_type), pointer dyed_obc_tracer::dyed_obc_tracer_cs::tr_reg => NULL() A pointer to the tracer registry.
-
real, dimension(:,:,:,:), pointer dyed_obc_tracer::dyed_obc_tracer_cs::tr => NULL() The array of tracers used in this subroutine, in g m-3?
-
integer, dimension(:), allocatable dyed_obc_tracer::dyed_obc_tracer_cs::ind_tr Indices returned by aof_set_coupler_flux if it is used and the surface tracer concentrations are to be provided to the coupler.
-
type(diag_ctrl), pointer dyed_obc_tracer::dyed_obc_tracer_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
type(mom_restart_cs), pointer dyed_obc_tracer::dyed_obc_tracer_cs::restart_csp => NULL() A pointer to the restart control structure.
-
type(vardesc), dimension(:), allocatable dyed_obc_tracer::dyed_obc_tracer_cs::tr_desc Descriptions and metadata for the tracers.
-
integer
- type
Describes the horizontal ocean grid with only dynamic memory arrays.
Public Members
-
type(mom_domain_type), pointer mom_dyn_horgrid::dyn_horgrid_type::domain => NULL() Ocean model domain.
-
type(mom_domain_type), pointer mom_dyn_horgrid::dyn_horgrid_type::domain_aux => NULL() A non-symmetric auxiliary domain type.
-
type(hor_index_type) mom_dyn_horgrid::dyn_horgrid_type::hi Horizontal index ranges.
-
integer
isc¶ The start i-index of cell centers within the computational domain.
-
integer
iec¶ The end i-index of cell centers within the computational domain.
-
integer
jsc¶ The start j-index of cell centers within the computational domain.
-
integer
jec¶ The end j-index of cell centers within the computational domain.
-
integer
isd¶ The start i-index of cell centers within the data domain.
-
integer
ied¶ The end i-index of cell centers within the data domain.
-
integer
jsd¶ The start j-index of cell centers within the data domain.
-
integer
jed¶ The end j-index of cell centers within the data domain.
-
integer
isg¶ The start i-index of cell centers within the global domain.
-
integer
ieg¶ The end i-index of cell centers within the global domain.
-
integer
jsg¶ The start j-index of cell centers within the global domain.
-
integer
jeg¶ The end j-index of cell centers within the global domain.
-
integer
iscb¶ The start i-index of cell vertices within the computational domain.
-
integer
iecb¶ The end i-index of cell vertices within the computational domain.
-
integer
jscb¶ The start j-index of cell vertices within the computational domain.
-
integer
jecb¶ The end j-index of cell vertices within the computational domain.
-
integer
isdb¶ The start i-index of cell vertices within the data domain.
-
integer
iedb¶ The end i-index of cell vertices within the data domain.
-
integer
jsdb¶ The start j-index of cell vertices within the data domain.
-
integer
jedb¶ The end j-index of cell vertices within the data domain.
-
integer
isgb¶ The start i-index of cell vertices within the global domain.
-
integer
iegb¶ The end i-index of cell vertices within the global domain.
-
integer
jsgb¶ The start j-index of cell vertices within the global domain.
-
integer
jegb¶ The end j-index of cell vertices within the global domain.
-
integer
isd_global¶ The value of isd in the global index space (decompoistion invariant).
-
integer
jsd_global¶ The value of isd in the global index space (decompoistion invariant).
-
integer
idg_offset¶ The offset between the corresponding global and local i-indices.
-
integer
jdg_offset¶ The offset between the corresponding global and local j-indices.
-
logical
symmetric¶ True if symmetric memory is used.
-
logical
nonblocking_updates¶ If true, non-blocking halo updates are allowed. The default is .false. (for now).
-
integer
first_direction¶ An integer that indicates which direction is to be updated first in directionally split parts of the calculation. This can be altered during the course of the run via calls to set_first_direction.
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::mask2dt 0 for land points and 1 for ocean points on the h-grid [nondim].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::geolatt The geographic latitude at q points [degrees of latitude] or [m].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::geolont The geographic longitude at q points [degrees of longitude] or [m].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::dxt dxT is delta x at h points [m].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::idxt 1/dxT [m-1].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::dyt dyT is delta y at h points [m].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::idyt IdyT is 1/dyT [m-1].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::areat The area of an h-cell [m2].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::iareat 1/areaT [m-2].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::sin_rot The sine of the angular rotation between the local model grid’s northward and the true northward directions [nondim].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::cos_rot The cosine of the angular rotation between the local model grid’s northward and the true northward directions [nondim].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::mask2dcu 0 for boundary points and 1 for ocean points on the u grid [nondim].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::geolatcu The geographic latitude at u points [degrees of latitude] or [m].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::geoloncu The geographic longitude at u points [degrees of longitude] or [m].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::dxcu dxCu is delta x at u points [m].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::idxcu 1/dxCu [m-1].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::dycu dyCu is delta y at u points [m].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::idycu 1/dyCu [m-1].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::dy_cu The unblocked lengths of the u-faces of the h-cell [m].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::iareacu The masked inverse areas of u-grid cells [m2].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::areacu The areas of the u-grid cells [m2].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::mask2dcv 0 for boundary points and 1 for ocean points on the v grid [nondim].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::geolatcv The geographic latitude at v points [degrees of latitude] or [m].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::geoloncv The geographic longitude at v points [degrees of longitude] or [m].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::dxcv dxCv is delta x at v points [m].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::idxcv 1/dxCv [m-1].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::dycv dyCv is delta y at v points [m].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::idycv 1/dyCv [m-1].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::dx_cv The unblocked lengths of the v-faces of the h-cell [m].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::iareacv The masked inverse areas of v-grid cells [m2].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::areacv The areas of the v-grid cells [m2].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::mask2dbu 0 for boundary points and 1 for ocean points on the q grid [nondim].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::geolatbu The geographic latitude at q points [degrees of latitude] or [m].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::geolonbu The geographic longitude at q points [degrees of longitude] or [m].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::dxbu dxBu is delta x at q points [m].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::idxbu 1/dxBu [m-1].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::dybu dyBu is delta y at q points [m].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::idybu 1/dyBu [m-1].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::areabu areaBu is the area of a q-cell [m2]
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::iareabu IareaBu = 1/areaBu [m-2].
-
real, dimension(:), pointer mom_dyn_horgrid::dyn_horgrid_type::gridlatt => NULL() The latitude of T points for the purpose of labeling the output axes. On many grids this is the same as geoLatT.
-
real, dimension(:), pointer mom_dyn_horgrid::dyn_horgrid_type::gridlatb => NULL() The latitude of B points for the purpose of labeling the output axes. On many grids this is the same as geoLatBu.
-
real, dimension(:), pointer mom_dyn_horgrid::dyn_horgrid_type::gridlont => NULL() The longitude of T points for the purpose of labeling the output axes. On many grids this is the same as geoLonT.
-
real, dimension(:), pointer mom_dyn_horgrid::dyn_horgrid_type::gridlonb => NULL() The longitude of B points for the purpose of labeling the output axes. On many grids this is the same as geoLonBu.
-
character(len=40) mom_dyn_horgrid::dyn_horgrid_type::x_axis_units The units that are used in labeling the x coordinate axes.
-
character(len=40) mom_dyn_horgrid::dyn_horgrid_type::y_axis_units The units that are used in labeling the y coordinate axes.
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::bathyt Ocean bottom depth at tracer points, in depth units [Z ~> m].
-
logical
bathymetry_at_vel¶ If true, there are separate values for the basin depths at velocity points. Otherwise the effects of of topography are entirely determined from thickness points.
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::dblock_u Topographic depths at u-points at which the flow is blocked [Z ~> m].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::dopen_u Topographic depths at u-points at which the flow is open at width dy_Cu [Z ~> m].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::dblock_v Topographic depths at v-points at which the flow is blocked [Z ~> m].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::dopen_v Topographic depths at v-points at which the flow is open at width dx_Cv [Z ~> m].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::coriolisbu The Coriolis parameter at corner points [T-1 ~> s-1].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::df_dx Derivative d/dx f (Coriolis parameter) at h-points [T-1 m-1 ~> s-1 m-1].
-
real, dimension(:,:), allocatable mom_dyn_horgrid::dyn_horgrid_type::df_dy Derivative d/dy f (Coriolis parameter) at h-points [T-1 m-1 ~> s-1 m-1].
-
real
areat_global¶ Global sum of h-cell area [m2].
-
real
iareat_global¶ Global sum of inverse h-cell area (1/areaT_global) [m-2].
-
real
south_lat¶ The latitude (or y-coordinate) of the first v-line.
-
real
west_lon¶ The longitude (or x-coordinate) of the first u-line.
-
real
len_lat= 0.¶ The latitudinal (or y-coord) extent of physical domain.
-
real
len_lon= 0.¶ The longitudinal (or x-coord) extent of physical domain.
-
real
rad_earth= 6.378e6¶ The radius of the planet [m].
-
real
max_depth¶ The maximum depth of the ocean [Z ~> m].
-
- type
The Extended Fixed Point (EFP) type provides a public interface for doing sums and taking differences with this type.
The use of this type is documented in Hallberg, R. & A. Adcroft, 2014: An Order-invariant Real-to-Integer Conversion Sum. Parallel Computing, 40(5-6), doi:10.1016/j.parco.2014.04.007.
Public Members
-
integer(kind=8), dimension(ni) mom_coms::efp_type::v The value in this type.
-
- type
This control structure holds parameters for the MOM_energetic_PBL module.
Unnamed Group
-
integer
id_ml_depth= -1¶ Diagnostic IDs.
-
integer
id_tke_wind= -1¶ Diagnostic IDs.
-
integer
id_tke_mixing= -1¶ Diagnostic IDs.
-
integer
id_tke_mke= -1¶ Diagnostic IDs.
-
integer
id_tke_conv= -1¶ Diagnostic IDs.
-
integer
id_tke_forcing= -1¶ Diagnostic IDs.
-
integer
id_tke_mech_decay= -1¶ Diagnostic IDs.
-
integer
id_tke_conv_decay= -1¶ Diagnostic IDs.
-
integer
id_mixing_length= -1¶ Diagnostic IDs.
-
integer
id_velocity_scale= -1¶ Diagnostic IDs.
-
integer
id_mstar_mix= -1¶ Diagnostic IDs.
-
integer
id_la_mod= -1¶ Diagnostic IDs.
-
integer
id_la= -1¶ Diagnostic IDs.
-
integer
id_mstar_lt= -1¶ Diagnostic IDs.
Public Members
-
real
vonkar= 0.41¶ The von Karman coefficient. This should be runtime, but because it is runtime in KPP and set to 0.4 it might change answers.
-
real
omega¶ The Earth’s rotation rate [T-1].
-
real
omega_frac¶ When setting the decay scale for turbulence, use this fraction of the absolute rotation rate blended with the local value of f, as sqrt((1-of)*f^2 + of*4*omega^2) [nondim].
-
real
nstar¶ The fraction of the TKE input to the mixed layer available to drive entrainment [nondim]. This quantity is the vertically integrated buoyancy production minus the vertically integrated dissipation of TKE produced by buoyancy.
-
logical
use_mld_iteration= .false.¶ False to use old ePBL method.
-
logical
mld_iteration_guess= .false.¶ False to default to guessing half the ocean depth for the iteration.
-
integer
max_mld_its¶ The maximum number of iterations that can be used to find a self-consistent mixed layer depth with Use_MLD_iteration.
-
real
mixlenexponent¶ Exponent in the mixing length shape-function. 1 is law-of-the-wall at top and bottom, 2 is more KPP like.
-
real
mke_to_tke_effic¶ The efficiency with which mean kinetic energy released by mechanically forced entrainment of the mixed layer is converted to TKE [nondim].
-
real
ustar_min¶ A minimum value of ustar to avoid numerical problems [Z T-1 ~> m s-1]. If the value is small enough, this should not affect the solution.
-
real
ekman_scale_coef¶ A nondimensional scaling factor controlling the inhibition of the diffusive length scale by rotation. Making this larger decreases the diffusivity in the planetary boundary layer.
-
real
translay_scale¶ A scale for the mixing length in the transition layer at the edge of the boundary layer as a fraction of the boundary layer thickness. The default is 0, but a value of 0.1 might be better justified by observations.
-
real
mld_tol¶ A tolerance for determining the boundary layer thickness when Use_MLD_iteration is true [Z ~> m].
-
real
min_mix_len¶ The minimum mixing length scale that will be used by ePBL [Z ~> m]. The default (0) does not set a minimum.
-
integer
wt_scheme¶ An enumerated value indicating the method for finding the turbulent velocity scale. There are currently two options: wT_mwT_from_cRoot_TKE is the original (TKE_remaining)^1/3 wT_from_RH18 is the version described by Reichl and Hallberg, 2018.
-
real
wstar_ustar_coef¶ A ratio relating the efficiency with which convectively released energy is converted to a turbulent velocity, relative to mechanically forced turbulent kinetic energy [nondim]. Making this larger increases the diffusivity.
-
real
vstar_surf_fac¶ If (wT_scheme == wT_from_RH18) this is the proportionality coefficient between ustar and the surface mechanical contribution to vstar [nondim].
-
real
vstar_scale_fac¶ An overall nondimensional scaling factor for vstar times a unit conversion factor [Z s T-1 m-1 ~> nondim]. Making this larger increases the diffusivity.
-
integer
mstar_scheme¶ An encoded integer to determine which formula is used to set mstar.
-
logical
mstar_flatcap= .true.¶ Set false to use asymptotic mstar cap.
-
real
mstar_cap¶ Since MSTAR is restoring undissipated energy to mixing, there must be a cap on how large it can be. This is definitely a function of latitude (Ekman limit), but will be taken as constant for now.
-
real
tke_decay¶ The ratio of the natural Ekman depth to the TKE decay scale [nondim].
-
real
fixed_mstar¶ Mstar is the ratio of the friction velocity cubed to the TKE available to drive entrainment, nondimensional. This quantity is the vertically integrated shear production minus the vertically integrated dissipation of TKE produced by shear. This value is used if the option for using a fixed mstar is used.
-
real
c_ek= 0.17¶ MSTAR Coefficient in rotation limit for mstar_scheme=OM4.
-
real
mstar_coef= 0.3¶ MSTAR coefficient in rotation/stabilizing balance for mstar_scheme=OM4.
-
real
rh18_mstar_cn1¶ MSTAR_N coefficient 1 (outter-most coefficient for fit). Value of 0.275 in RH18. Increasing this coefficient increases mechanical mixing for all values of Hf/ust, but is most effective at low values (weakly developed OSBLs).
-
real
rh18_mstar_cn2¶ MSTAR_N coefficient 2 (coefficient outside of exponential decay). Value of 8.0 in RH18. Increasing this coefficient increases MSTAR for all values of HF/ust, with a consistent affect across a wide range of Hf/ust.
-
real
rh18_mstar_cn3¶ MSTAR_N coefficient 3 (exponential decay coefficient). Value of -5.0 in RH18. Increasing this increases how quickly the value of MSTAR decreases as Hf/ust increases.
-
real
rh18_mstar_cs1¶ MSTAR_S coefficient for RH18 in stabilizing limit. Value of 0.2 in RH18.
-
real
rh18_mstar_cs2¶ MSTAR_S exponent for RH18 in stabilizing limit. Value of 0.4 in RH18.
-
real
mstar_convect_coef¶ Factor to reduce mstar when statically unstable.
-
logical
use_lt= .false.¶ Flag for using LT in Energy calculation.
-
integer
lt_enhance_form¶ Integer for Enhancement functional form (various options)
-
real
lt_enhance_coef¶ Coefficient in fit for Langmuir Enhancment.
-
real
lt_enhance_exp¶ Exponent in fit for Langmuir Enhancement.
-
real
lac_mldoek¶ Coefficient for Langmuir number modification based on the ratio of the mixed layer depth over the Ekman depth.
-
real
lac_mldoob_stab¶ Coefficient for Langmuir number modification based on the ratio of the mixed layer depth over the Obukov depth with stablizing forcing.
-
real
lac_ekoob_stab¶ Coefficient for Langmuir number modification based on the ratio of the Ekman depth over the Obukov depth with stablizing forcing.
-
real
lac_mldoob_un¶ Coefficient for Langmuir number modification based on the ratio of the mixed layer depth over the Obukov depth with destablizing forcing.
-
real
lac_ekoob_un¶ Coefficient for Langmuir number modification based on the ratio of the Ekman depth over the Obukov depth with destablizing forcing.
-
real
max_enhance_m= 5.¶ The maximum allowed LT enhancement to the mixing.
-
type(time_type), pointer mom_energetic_pbl::energetic_pbl_cs::time =>NULL() A pointer to the ocean model’s clock.
-
logical
tke_diagnostics= .false.¶ If true, diagnostics of the TKE budget are being calculated.
-
logical
answers_2018¶ If true, use the order of arithmetic and expressions that recover the answers from the end of 2018. Otherwise, use updated and more robust forms of the same expressions.
-
logical
orig_pe_calc¶ If true, the ePBL code uses the original form of the potential energy change code. Otherwise, it uses a newer version that can work with successive increments to the diffusivity in upward or downward passes.
-
type(diag_ctrl), pointer mom_energetic_pbl::energetic_pbl_cs::diag =>NULL() A structure that is used to regulate the timing of diagnostic output.
-
real, dimension(:,:), allocatable mom_energetic_pbl::energetic_pbl_cs::ml_depth The mixed layer depth determined by active mixing in ePBL [Z ~> m].
-
real, dimension(:,:), allocatable mom_energetic_pbl::energetic_pbl_cs::diag_tke_wind The wind source of TKE [kg m-3 Z3 T-3 ~> W m-2].
-
real, dimension(:,:), allocatable mom_energetic_pbl::energetic_pbl_cs::diag_tke_mke The resolved KE source of TKE [kg m-3 Z3 T-3 ~> W m-2].
-
real, dimension(:,:), allocatable mom_energetic_pbl::energetic_pbl_cs::diag_tke_conv The convective source of TKE [kg m-3 Z3 T-3 ~> W m-2].
-
real, dimension(:,:), allocatable mom_energetic_pbl::energetic_pbl_cs::diag_tke_forcing The TKE sink required to mix surface penetrating shortwave heating.
-
real, dimension(:,:), allocatable mom_energetic_pbl::energetic_pbl_cs::diag_tke_mech_decay The decay of mechanical TKE [kg m-3 Z3 T-3 ~> W m-2].
-
real, dimension(:,:), allocatable mom_energetic_pbl::energetic_pbl_cs::diag_tke_conv_decay The decay of convective TKE [kg m-3 Z3 T-3 ~> W m-2].
-
real, dimension(:,:), allocatable mom_energetic_pbl::energetic_pbl_cs::diag_tke_mixing The work done by TKE to deepen the mixed layer [kg m-3 Z3 T-3 ~> W m-2].
-
real, dimension(:,:), allocatable mom_energetic_pbl::energetic_pbl_cs::mstar_mix Mstar used in EPBL [nondim].
-
real, dimension(:,:), allocatable mom_energetic_pbl::energetic_pbl_cs::mstar_lt Mstar due to Langmuir turbulence [nondim].
-
real, dimension(:,:), allocatable mom_energetic_pbl::energetic_pbl_cs::la Langmuir number [nondim].
-
real, dimension(:,:), allocatable mom_energetic_pbl::energetic_pbl_cs::la_mod Modified Langmuir number [nondim].
-
real, dimension(:,:,:), allocatable mom_energetic_pbl::energetic_pbl_cs::velocity_scale The velocity scale used in getting Kd [Z T-1 ~> m s-1].
-
real, dimension(:,:,:), allocatable mom_energetic_pbl::energetic_pbl_cs::mixing_length The length scale used in getting Kd [Z ~> m].
-
integer
- type
The control structure holding parametes for the MOM_entrain_diffusive module.
Public Members
-
logical
bulkmixedlayer¶ If true, a refined bulk mixed layer is used with GVnk_rho_varies variable density mixed & buffer layers.
-
logical
correct_density¶ If true, the layer densities are restored toward their target variables by the diapycnal mixing.
-
integer
max_ent_it¶ The maximum number of iterations that may be used to calculate the diapycnal entrainment.
-
real
tolerance_ent¶ The tolerance with which to solve for entrainment values [H ~> m or kg m-2].
-
type(diag_ctrl), pointer mom_entrain_diffusive::entrain_diffusive_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
integer
id_kd= -1¶ Diagnostic ID for diffusivity.
-
integer
id_diff_work= -1¶ Diagnostic ID for mixing work.
-
logical
- type
A control structure for the equation of state.
Public Members
-
integer
form_of_eos= 0¶ The equation of state to use.
-
integer
form_of_tfreeze= 0¶ The expression for the potential temperature of the freezing point.
-
logical
eos_quadrature¶ If true, always use the generic (quadrature) code for the integrals of density.
-
logical
compressible= .true.¶ If true, in situ density is a function of pressure.
-
real
rho_t0_s0¶ The density at T=0, S=0 [kg m-3].
-
real
drho_dt¶ The partial derivative of density with temperature [kg m-3 degC-1].
-
real
drho_ds¶ The partial derivative of density with salinity [kg m-3 ppt-1].
-
real
tfr_s0_p0¶ The freezing potential temperature at S=0, P=0 [degC].
-
real
dtfr_ds¶ The derivative of freezing point with salinity [degC ppt-1].
-
real
dtfr_dp¶ The derivative of freezing point with pressure [degC Pa-1].
-
integer
- type
A type for conveniently passing around ePBL diagnostics for a column.
Unnamed Group
-
real
dtke_conv¶ Local column copies of energy change diagnostics, all in [kg m-3 Z3 T-3 ~> W m-2].
-
real
dtke_forcing¶ Local column copies of energy change diagnostics, all in [kg m-3 Z3 T-3 ~> W m-2].
-
real
dtke_wind¶ Local column copies of energy change diagnostics, all in [kg m-3 Z3 T-3 ~> W m-2].
-
real
dtke_mixing¶ Local column copies of energy change diagnostics, all in [kg m-3 Z3 T-3 ~> W m-2].
-
real
dtke_mke¶ Local column copies of energy change diagnostics, all in [kg m-3 Z3 T-3 ~> W m-2].
-
real
dtke_mech_decay¶ Local column copies of energy change diagnostics, all in [kg m-3 Z3 T-3 ~> W m-2].
-
real
dtke_conv_decay¶ Local column copies of energy change diagnostics, all in [kg m-3 Z3 T-3 ~> W m-2].
-
real
la¶ The value of the Langmuir number [nondim].
-
real
lamod¶ The modified Langmuir number by convection [nondim].
-
real
mstar¶ The value of mstar used in ePBL [nondim].
-
real
mstar_lt¶ The portion of mstar due to Langmuir turbulence [nondim].
-
real, dimension(:), allocatable mom_energetic_pbl::epbl_column_diags::dt_expect Expected temperature changes [degC].
-
real, dimension(:), allocatable mom_energetic_pbl::epbl_column_diags::ds_expect Expected salinity changes [ppt].
-
real
- type
A structure with information about a single restart field.
Private Members
-
type(vardesc) mom_restart::field_restart::vars Description of a field that is to be read from or written to the restart file.
-
logical
mand_var¶ If .true. the run will abort if this field is not successfully read from the restart file.
-
logical
initialized¶ .true. if this field has been read from the restart file.
-
character(len=32) mom_restart::field_restart::var_name A name by which a variable may be queried.
-
- type
The valid lines extracted from an input parameter file without comments.
Unnamed Group
-
integer
num_lines= 0¶ The number of lines in this type.
-
character(len=input_str_length), dimension(:), pointer mom_file_parser::file_data_type::line => NULL() The line content.
-
logical, dimension(:), pointer mom_file_parser::file_data_type::line_used => NULL() If true, the line has been read.
-
integer
-
interface
file_exists¶ Indicate whether a file exists, perhaps with domain decomposition.
Private Functions
-
logical function mom_io::file_exists::fms_file_exists(filename filename, domain domain, no_domain no_domain) Returns true if the named file or its domain-decomposed variant exists.
- Parameters
[in] filename: The name of the file being inquired about[in] domain: The mpp domain2d that describes the decomposition[in] no_domain: This file does not use domain decomposition
-
logical function mom_io::file_exists::mom_file_exists(filename filename, MOM_Domain MOM_Domain) Returns true if the named file or its domain-decomposed variant exists.
- Parameters
[in] filename: The name of the file being inquired about[in] mom_domain: The MOM_Domain that describes the decomposition
-
- type
Control structure for open boundaries that read from files. Probably lots to update here.
Public Members
-
real
tide_flow= 3.0e6¶ Placeholder for now…
-
real
-
interface
fill_boundaries¶ Fill grid edges.
Private Functions
-
real function, dimension(0:size(m,1)+1,0:size(m,2)+1) mom_horizontal_regridding::fill_boundaries::fill_boundaries_real(m m, cyclic_x cyclic_x, tripolar_n tripolar_n) Fill grid edges for real data.
- Parameters
[in] m: input array (ND)[in] cyclic_x: True if domain is zonally re-entrant[in] tripolar_n: True if domain has an Arctic fold
-
integer function, dimension(0:size(m,1)+1,0:size(m,2)+1) mom_horizontal_regridding::fill_boundaries::fill_boundaries_int(m m, cyclic_x cyclic_x, tripolar_n tripolar_n) Fill grid edges for integer data.
- Parameters
[in] m: input array (ND)[in] cyclic_x: True if domain is zonally re-entrant[in] tripolar_n: True if domain has an Arctic fold
-
-
interface
fill_boundaries¶ Fill grid edges.
Private Functions
-
real function, dimension(0:size(m,1)+1,0:size(m,2)+1) midas_vertmap::fill_boundaries::fill_boundaries_real(m m, cyclic_x cyclic_x, tripolar_n tripolar_n) fill grid edges
- Return
output filled array
- Parameters
[in] m: input array[in] cyclic_x: zonal cyclic condition[in] tripolar_n: northern fold condition
-
integer function, dimension(0:size(m,1)+1,0:size(m,2)+1) midas_vertmap::fill_boundaries::fill_boundaries_int(m m, cyclic_x cyclic_x, tripolar_n tripolar_n) Fill grid edges.
- Return
output filled array
- Parameters
[in] m: input array[in] cyclic_x: zonal cyclic condition[in] tripolar_n: northern fold condition
-
-
interface
fill_symmetric_edges¶ Do a set of halo updates that fill in the values at the duplicated edges of a staggered symmetric memory domain.
Private Functions
-
subroutine
fill_vector_symmetric_edges_2d(u_cmpt u_cmpt, v_cmpt v_cmpt, MOM_dom MOM_dom, stagger stagger, scalar scalar, clock clock)¶ fill_vector_symmetric_edges_2d does an usual set of halo updates that only fill in the values at the edge of a pair of symmetric memory two-dimensional arrays representing the compontents of a two-dimensional horizontal vector. If symmetric memory is not being used, this subroutine does nothing except to possibly turn optional cpu clocks on or off.
- Parameters
[inout] u_cmpt: The nominal zonal (u) component of the vector pair which is having its halos points exchanged.[inout] v_cmpt: The nominal meridional (v) component of the vector pair which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] stagger: An optional flag, which may be one of A_GRID, BGRID_NE, or CGRID_NE, indicating where the two components of the vector are discretized. Omitting stagger is the same as setting it to CGRID_NE.[in] scalar: An optional argument indicating whether.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
-
interface
find_eta¶ Calculates the heights of sruface or all interfaces from layer thicknesses.
Private Functions
-
subroutine
find_eta_2d(h h, tv tv, G G, GV GV, US US, eta eta, eta_bt eta_bt, halo_size halo_size, eta_to_m eta_to_m)¶ Calculates the free surface height, using the appropriate form for consistency with the calculation of the pressure gradient forces. Additionally, the sea surface height may be adjusted for consistency with the corresponding time-average quantity from the barotropic calculation.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] h: Layer thicknesses [H ~> m or kg m-2][in] tv: A structure pointing to various thermodynamic variables.[out] eta: free surface height relative to mean sea level (z=0) often [Z ~> m].[in] eta_bt: optional barotropic variable that gives the “correct” free surface height (Boussinesq) or total water column mass per unit area (non-Boussinesq) [H ~> m or kg m-2].[in] halo_size: width of halo points on which to calculate eta.[in] eta_to_m: The conversion factor from the units of eta to m; by default this is USZ_to_m.
-
subroutine
find_eta_3d(h h, tv tv, G G, GV GV, US US, eta eta, eta_bt eta_bt, halo_size halo_size, eta_to_m eta_to_m)¶ Calculates the heights of all interfaces between layers, using the appropriate form for consistency with the calculation of the pressure gradient forces. Additionally, these height may be dilated for consistency with the corresponding time-average quantity from the barotropic calculation.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] h: Layer thicknesses [H ~> m or kg m-2][in] tv: A structure pointing to various thermodynamic variables.[out] eta: layer interface heights [Z ~> m] or 1/eta_to_m m).[in] eta_bt: optional barotropic variable that gives the “correct” free surface height (Boussinesq) or total water column mass per unit area (non-Boussinesq). This is used to dilate the layer. thicknesses when calculating interfaceheights [H ~> m or kg m-2].[in] halo_size: width of halo points on which to calculate eta.[in] eta_to_m: The conversion factor from the units of eta to m; by default this is USZ_to_m.
-
subroutine
- type
Structure that contains pointers to the boundary forcing used to drive the liquid ocean simulated by MOM.
Data in this type is allocated in the module MOM_surface_forcing.F90, of which there are three: solo, coupled, and ice-shelf. Alternatively, they are allocated in MESO_surface_forcing.F90, which is a special case of solo_driver/MOM_surface_forcing.F90.
Public Members
-
real, dimension(:,:), pointer mom_forcing_type::forcing::ustar => NULL() surface friction velocity scale [Z T-1 ~> m s-1].
-
real, dimension(:,:), pointer mom_forcing_type::forcing::ustar_gustless => NULL() surface friction velocity scale without any
-
real, dimension(:,:), pointer mom_forcing_type::forcing::buoy => NULL() buoyancy flux [L2 T-3 ~> m2 s-3]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::sw => NULL() shortwave [W m-2]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::sw_vis_dir => NULL() visible, direct shortwave [W m-2]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::sw_vis_dif => NULL() visible, diffuse shortwave [W m-2]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::sw_nir_dir => NULL() near-IR, direct shortwave [W m-2]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::sw_nir_dif => NULL() near-IR, diffuse shortwave [W m-2]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::lw => NULL() longwave [W m-2] (typically negative)
-
real, dimension(:,:), pointer mom_forcing_type::forcing::latent => NULL() latent [W m-2] (typically < 0)
-
real, dimension(:,:), pointer mom_forcing_type::forcing::sens => NULL() sensible [W m-2] (typically negative)
-
real, dimension(:,:), pointer mom_forcing_type::forcing::seaice_melt_heat => NULL() sea ice and snow melt or formation [W m-2] (typically negative)
-
real, dimension(:,:), pointer mom_forcing_type::forcing::heat_added => NULL() additional heat flux from SST restoring or flux adjustments [W m-2]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::latent_evap_diag => NULL() latent [W m-2] from evaporating liquid water (typically < 0)
-
real, dimension(:,:), pointer mom_forcing_type::forcing::latent_fprec_diag => NULL() latent [W m-2] from melting fprec (typically < 0)
-
real, dimension(:,:), pointer mom_forcing_type::forcing::latent_frunoff_diag => NULL() latent [W m-2] from melting frunoff (calving) (typically < 0)
-
real, dimension(:,:), pointer mom_forcing_type::forcing::evap => NULL() (-1)*fresh water flux evaporated out of the ocean [kg m-2 s-1]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::lprec => NULL() precipitating liquid water into the ocean [kg m-2 s-1]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::fprec => NULL() precipitating frozen water into the ocean [kg m-2 s-1]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::vprec => NULL() virtual liquid precip associated w/ SSS restoring [kg m-2 s-1]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::lrunoff => NULL() liquid river runoff entering ocean [kg m-2 s-1]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::frunoff => NULL() frozen river runoff (calving) entering ocean [kg m-2 s-1]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::seaice_melt => NULL() snow/seaice melt (positive) or formation (negative) [kg m-2 s-1]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::netmassin => NULL() Sum of water mass flux out of the ocean [kg m-2 s-1].
-
real, dimension(:,:), pointer mom_forcing_type::forcing::netmassout => NULL() Net water mass flux into of the ocean [kg m-2 s-1].
-
real, dimension(:,:), pointer mom_forcing_type::forcing::netsalt => NULL() Net salt entering the ocean [kgSalt m-2 s-1].
-
real, dimension(:,:), pointer mom_forcing_type::forcing::heat_content_cond => NULL() heat content associated with condensating water [W m-2]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::heat_content_lprec => NULL() heat content associated with liquid >0 precip [W m-2] (diagnostic)
-
real, dimension(:,:), pointer mom_forcing_type::forcing::heat_content_icemelt => NULL() heat content associated with snow/seaice melt/formation [W/m^2]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::heat_content_fprec => NULL() heat content associated with frozen precip [W m-2]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::heat_content_vprec => NULL() heat content associated with virtual >0 precip [W m-2]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::heat_content_lrunoff => NULL() heat content associated with liquid runoff [W m-2]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::heat_content_frunoff => NULL() heat content associated with frozen runoff [W m-2]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::heat_content_massout => NULL() heat content associated with mass leaving ocean [W m-2]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::heat_content_massin => NULL() heat content associated with mass entering ocean [W m-2]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::salt_flux => NULL() net salt flux into the ocean [kgSalt m-2 s-1]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::salt_flux_in => NULL() salt flux provided to the ocean from coupler [kgSalt m-2 s-1]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::salt_flux_added => NULL() additional salt flux from restoring or flux adjustment before adjustment
-
real, dimension(:,:), pointer mom_forcing_type::forcing::p_surf_full => NULL() Pressure at the top ocean interface [Pa]. if there is sea-ice, then p_surf_flux is at ice-ocean interface.
-
real, dimension(:,:), pointer mom_forcing_type::forcing::p_surf => NULL() Pressure at the top ocean interface [Pa] as used to drive the ocean model. If p_surf is limited, p_surf may be smaller than p_surf_full, otherwise they are the same.
-
real, dimension(:,:), pointer mom_forcing_type::forcing::p_surf_ssh => NULL() Pressure at the top ocean interface [Pa] that is used in corrections to the sea surface height field that is passed back to the calling routines. p_surf_SSH may point to p_surf or to p_surf_full.
-
logical
accumulate_p_surf= .false.¶ If true, the surface pressure due to the atmosphere and various types of ice needs to be accumulated, and the surface pressure explicitly reset to zero at the driver level when appropriate.
-
real, dimension(:,:), pointer mom_forcing_type::forcing::tke_tidal => NULL() tidal energy source driving mixing in bottom boundary layer [W m-2]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::ustar_tidal => NULL() tidal contribution to bottom ustar [Z T-1 ~> m s-1]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::ustar_berg => NULL() iceberg contribution to top ustar [Z T-1 ~> m s-1].
-
real, dimension(:,:), pointer mom_forcing_type::forcing::area_berg => NULL() area of ocean surface covered by icebergs [m2 m-2]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::mass_berg => NULL() mass of icebergs [kg m-2]
-
real, dimension(:,:), pointer mom_forcing_type::forcing::ustar_shelf => NULL() Friction velocity under ice-shelves [Z T-1 ~> m s-1]. as computed by the ocean at the previous time step.
-
real, dimension(:,:), pointer mom_forcing_type::forcing::frac_shelf_h => NULL() Fractional ice shelf coverage of h-cells, nondimensional from 0 to 1. This is only associated if ice shelves are enabled, and are exactly 0 away from shelves or on land.
-
real, dimension(:,:), pointer mom_forcing_type::forcing::iceshelf_melt => NULL() Ice shelf melt rate (positive) or freezing (negative) [m year-1].
-
real
vprecglobaladj¶ adjustment to restoring vprec to zero out global net [kg m-2 s-1]
-
real
saltfluxglobaladj¶ adjustment to restoring salt flux to zero out global net [kgSalt m-2 s-1]
-
real
netfwglobaladj¶ adjustment to net fresh water to zero out global net [kg m-2 s-1]
-
real
vprecglobalscl¶ scaling of restoring vprec to zero out global net ( -1..1 ) [nondim]
-
real
saltfluxglobalscl¶ scaling of restoring salt flux to zero out global net ( -1..1 ) [nondim]
-
real
netfwglobalscl¶ scaling of net fresh water to zero out global net ( -1..1 ) [nondim]
-
logical
fluxes_used= .true.¶ If true, all of the heat, salt, and mass fluxes have been applied to the ocean.
-
real
dt_buoy_accum= -1.0¶ The amount of time over which the buoyancy fluxes should be applied [s]. If negative, this forcing type variable has not yet been inialized.
-
real
c_p¶ heat capacity of seawater [J kg-1 degC-1]. C_p is is the same value as in thermovar_ptrs_type.
-
type(coupler_2d_bc_type) mom_forcing_type::forcing::tr_fluxes This structure contains arrays of of named fields used for passive tracer fluxes. All arrays in tr_fluxes use the coupler indexing, which has no halos. This is not a convenient convention, but imposed on MOM6 by the coupler.
-
integer
num_msg= 0¶ Number of messages issued about excessive SW penetration.
-
integer
max_msg= 2¶ Maximum number of messages to issue about excessive SW penetration.
-
- type
Structure that defines the id handles for the forcing type.
Unnamed Group
-
integer
id_prcme= -1¶ Forcing diagnostic handles.
-
integer
id_evap= -1¶ Forcing diagnostic handles.
-
integer
id_precip= -1¶ Forcing diagnostic handles.
-
integer
id_vprec= -1¶ Forcing diagnostic handles.
-
integer
id_lprec= -1¶ Forcing diagnostic handles.
-
integer
id_fprec= -1¶ Forcing diagnostic handles.
-
integer
id_lrunoff= -1¶ Forcing diagnostic handles.
-
integer
id_frunoff= -1¶ Forcing diagnostic handles.
-
integer
id_net_massout= -1¶ Forcing diagnostic handles.
-
integer
id_net_massin= -1¶ Forcing diagnostic handles.
-
integer
id_massout_flux= -1¶ Forcing diagnostic handles.
-
integer
id_massin_flux= -1¶ Forcing diagnostic handles.
-
integer
id_seaice_melt= -1¶ Forcing diagnostic handles.
-
integer
id_total_prcme= -1¶ Forcing diagnostic handles.
-
integer
id_total_evap= -1¶ Forcing diagnostic handles.
-
integer
id_total_precip= -1¶ Forcing diagnostic handles.
-
integer
id_total_vprec= -1¶ Forcing diagnostic handles.
-
integer
id_total_lprec= -1¶ Forcing diagnostic handles.
-
integer
id_total_fprec= -1¶ Forcing diagnostic handles.
-
integer
id_total_lrunoff= -1¶ Forcing diagnostic handles.
-
integer
id_total_frunoff= -1¶ Forcing diagnostic handles.
-
integer
id_total_net_massout= -1¶ Forcing diagnostic handles.
-
integer
id_total_net_massin= -1¶ Forcing diagnostic handles.
-
integer
id_total_seaice_melt= -1¶ Forcing diagnostic handles.
-
integer
id_prcme_ga= -1¶ Forcing diagnostic handles.
-
integer
id_evap_ga= -1¶ Forcing diagnostic handles.
-
integer
id_lprec_ga= -1¶ Forcing diagnostic handles.
-
integer
id_fprec_ga= -1¶ Forcing diagnostic handles.
-
integer
id_precip_ga= -1¶ Forcing diagnostic handles.
-
integer
id_vprec_ga= -1¶ Forcing diagnostic handles.
-
integer
id_net_heat_coupler= -1¶ Forcing diagnostic handles.
-
integer
id_net_heat_surface= -1¶ Forcing diagnostic handles.
-
integer
id_sens= -1¶ Forcing diagnostic handles.
-
integer
id_lwlatsens= -1¶ Forcing diagnostic handles.
-
integer
id_sw= -1¶ Forcing diagnostic handles.
-
integer
id_lw= -1¶ Forcing diagnostic handles.
-
integer
id_sw_vis= -1¶ Forcing diagnostic handles.
-
integer
id_sw_nir= -1¶ Forcing diagnostic handles.
-
integer
id_lat_evap= -1¶ Forcing diagnostic handles.
-
integer
id_lat_frunoff= -1¶ Forcing diagnostic handles.
-
integer
id_lat= -1¶ Forcing diagnostic handles.
-
integer
id_lat_fprec= -1¶ Forcing diagnostic handles.
-
integer
id_heat_content_lrunoff= -1¶ Forcing diagnostic handles.
-
integer
id_heat_content_frunoff= -1¶ Forcing diagnostic handles.
-
integer
id_heat_content_lprec= -1¶ Forcing diagnostic handles.
-
integer
id_heat_content_fprec= -1¶ Forcing diagnostic handles.
-
integer
id_heat_content_cond= -1¶ Forcing diagnostic handles.
-
integer
id_heat_content_surfwater= -1¶ Forcing diagnostic handles.
-
integer
id_heat_content_vprec= -1¶ Forcing diagnostic handles.
-
integer
id_heat_content_massout= -1¶ Forcing diagnostic handles.
-
integer
id_heat_added= -1¶ Forcing diagnostic handles.
-
integer
id_heat_content_massin= -1¶ Forcing diagnostic handles.
-
integer
id_hfrainds= -1¶ Forcing diagnostic handles.
-
integer
id_hfrunoffds= -1¶ Forcing diagnostic handles.
-
integer
id_seaice_melt_heat= -1¶ Forcing diagnostic handles.
-
integer
id_heat_content_icemelt= -1¶ Forcing diagnostic handles.
-
integer
id_total_net_heat_coupler= -1¶ Forcing diagnostic handles.
-
integer
id_total_net_heat_surface= -1¶ Forcing diagnostic handles.
-
integer
id_total_sens= -1¶ Forcing diagnostic handles.
-
integer
id_total_lwlatsens= -1¶ Forcing diagnostic handles.
-
integer
id_total_sw= -1¶ Forcing diagnostic handles.
-
integer
id_total_lw= -1¶ Forcing diagnostic handles.
-
integer
id_total_lat_evap= -1¶ Forcing diagnostic handles.
-
integer
id_total_lat_frunoff= -1¶ Forcing diagnostic handles.
-
integer
id_total_lat= -1¶ Forcing diagnostic handles.
-
integer
id_total_lat_fprec= -1¶ Forcing diagnostic handles.
-
integer
id_total_heat_content_lrunoff= -1¶ Forcing diagnostic handles.
-
integer
id_total_heat_content_frunoff= -1¶ Forcing diagnostic handles.
-
integer
id_total_heat_content_lprec= -1¶ Forcing diagnostic handles.
-
integer
id_total_heat_content_fprec= -1¶ Forcing diagnostic handles.
-
integer
id_total_heat_content_cond= -1¶ Forcing diagnostic handles.
-
integer
id_total_heat_content_surfwater= -1¶ Forcing diagnostic handles.
-
integer
id_total_heat_content_vprec= -1¶ Forcing diagnostic handles.
-
integer
id_total_heat_content_massout= -1¶ Forcing diagnostic handles.
-
integer
id_total_heat_added= -1¶ Forcing diagnostic handles.
-
integer
id_total_heat_content_massin= -1¶ Forcing diagnostic handles.
-
integer
id_total_seaice_melt_heat= -1¶ Forcing diagnostic handles.
-
integer
id_total_heat_content_icemelt= -1¶ Forcing diagnostic handles.
-
integer
id_net_heat_coupler_ga= -1¶ Forcing diagnostic handles.
-
integer
id_net_heat_surface_ga= -1¶ Forcing diagnostic handles.
-
integer
id_sens_ga= -1¶ Forcing diagnostic handles.
-
integer
id_lwlatsens_ga= -1¶ Forcing diagnostic handles.
-
integer
id_sw_ga= -1¶ Forcing diagnostic handles.
-
integer
id_lw_ga= -1¶ Forcing diagnostic handles.
-
integer
id_lat_ga= -1¶ Forcing diagnostic handles.
-
integer
id_saltflux= -1¶ Forcing diagnostic handles.
-
integer
id_saltfluxin= -1¶ Forcing diagnostic handles.
-
integer
id_saltfluxadded= -1¶ Forcing diagnostic handles.
-
integer
id_total_saltflux= -1¶ Forcing diagnostic handles.
-
integer
id_total_saltfluxin= -1¶ Forcing diagnostic handles.
-
integer
id_total_saltfluxadded= -1¶ Forcing diagnostic handles.
-
integer
id_vprecglobaladj= -1¶ Forcing diagnostic handles.
-
integer
id_vprecglobalscl= -1¶ Forcing diagnostic handles.
-
integer
id_saltfluxglobaladj= -1¶ Forcing diagnostic handles.
-
integer
id_saltfluxglobalscl= -1¶ Forcing diagnostic handles.
-
integer
id_netfwglobaladj= -1¶ Forcing diagnostic handles.
-
integer
id_netfwglobalscl= -1¶ Forcing diagnostic handles.
-
integer
id_taux= -1¶ Forcing diagnostic handles.
-
integer
id_tauy= -1¶ Forcing diagnostic handles.
-
integer
id_ustar= -1¶ Forcing diagnostic handles.
-
integer
id_psurf= -1¶ Forcing diagnostic handles.
-
integer
id_tke_tidal= -1¶ Forcing diagnostic handles.
-
integer
id_buoy= -1¶ Forcing diagnostic handles.
-
integer
id_ustar_berg= -1¶ Forcing diagnostic handles.
-
integer
id_area_berg= -1¶ Forcing diagnostic handles.
-
integer
id_mass_berg= -1¶ Forcing diagnostic handles.
-
integer
id_ustar_ice_cover= -1¶ Forcing diagnostic handles.
-
integer
id_frac_ice_cover= -1¶ Forcing diagnostic handles.
-
integer
id_clock_forcing= -1¶ CPU clock id.
-
integer
- type
Control structure for geothermal heating.
Public Members
-
real
drcv_dt_inplace¶ The value of dRcv_dT above which (dRcv_dT is negative) the water is heated in place instead of moving upward between layers [kg m-3 degC-1].
-
real, dimension(:,:), pointer mom_geothermal::geothermal_cs::geo_heat => NULL() The geothermal heat flux [W m-2].
-
real
geothermal_thick¶ The thickness over which geothermal heating is applied [m] (not [H]).
-
logical
apply_geothermal¶ If true, geothermal heating will be applied otherwise GEOTHERMAL_SCALE has been set to 0 and there is no heat to apply.
-
type(time_type), pointer mom_geothermal::geothermal_cs::time => NULL() A pointer to the ocean model’s clock.
-
type(diag_ctrl), pointer mom_geothermal::geothermal_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
real
-
interface
get_param¶ An overloaded interface to read and log the values of various types of parameters.
Unnamed Group
-
subroutine mom_file_parser::get_param::get_param_int(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, fail_if_missing fail_if_missing, do_not_read do_not_read, do_not_log do_not_log, static_value static_value, layoutParam layoutParam, debuggingParam debuggingParam) This subroutine reads the value of an integer model parameter from a parameter file and logs it in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file and logged[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] static_value: If this parameter is static, it takes this value, which can be compared for consistency with what is in the parameter file.[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file[in] do_not_read: If present and true, do not read a value for this parameter, although it might be logged.[in] do_not_log: If present and true, do not log this parameter to the documentation files[in] layoutparam: If present and true, this parameter is logged in the layout parameter file[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file
-
subroutine mom_file_parser::get_param::get_param_real(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, fail_if_missing fail_if_missing, do_not_read do_not_read, do_not_log do_not_log, static_value static_value, debuggingParam debuggingParam, scale scale, unscaled unscaled) This subroutine reads the value of a real model parameter from a parameter file and logs it in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file and logged[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] static_value: If this parameter is static, it takes this value, which can be compared for consistency with what is in the parameter file.[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file[in] do_not_read: If present and true, do not read a value for this parameter, although it might be logged.[in] do_not_log: If present and true, do not log this parameter to the documentation files[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file[in] scale: A scaling factor that the parameter is multiplied by before it is returned.[out] unscaled: The value of the parameter that would be returned without any multiplication by a scaling factor.
-
subroutine mom_file_parser::get_param::get_param_logical(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, fail_if_missing fail_if_missing, do_not_read do_not_read, do_not_log do_not_log, static_value static_value, layoutParam layoutParam, debuggingParam debuggingParam) This subroutine reads the value of a logical model parameter from a parameter file and logs it in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file and logged[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] static_value: If this parameter is static, it takes this value, which can be compared for consistency with what is in the parameter file.[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file[in] do_not_read: If present and true, do not read a value for this parameter, although it might be logged.[in] do_not_log: If present and true, do not log this parameter to the documentation files[in] layoutparam: If present and true, this parameter is logged in the layout parameter file[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file
-
subroutine mom_file_parser::get_param::get_param_char(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, fail_if_missing fail_if_missing, do_not_read do_not_read, do_not_log do_not_log, static_value static_value, layoutParam layoutParam, debuggingParam debuggingParam) This subroutine reads the value of a character string model parameter from a parameter file and logs it in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file and logged[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] static_value: If this parameter is static, it takes this value, which can be compared for consistency with what is in the parameter file.[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file[in] do_not_read: If present and true, do not read a value for this parameter, although it might be logged.[in] do_not_log: If present and true, do not log this parameter to the documentation files[in] layoutparam: If present and true, this parameter is logged in the layout parameter file[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file
-
subroutine mom_file_parser::get_param::get_param_char_array(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, fail_if_missing fail_if_missing, do_not_read do_not_read, do_not_log do_not_log, static_value static_value) This subroutine reads the values of an array of character string model parameters from a parameter file and logs them in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file and logged[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] static_value: If this parameter is static, it takes this value, which can be compared for consistency with what is in the parameter file.[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file[in] do_not_read: If present and true, do not read a value for this parameter, although it might be logged.[in] do_not_log: If present and true, do not log this parameter to the documentation files
-
subroutine mom_file_parser::get_param::get_param_time(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, fail_if_missing fail_if_missing, do_not_read do_not_read, do_not_log do_not_log, timeunit timeunit, static_value static_value, layoutParam layoutParam, debuggingParam debuggingParam, log_as_date log_as_date) This subroutine reads the value of a time-type model parameter from a parameter file and logs it in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file and logged[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] static_value: If this parameter is static, it takes this value, which can be compared for consistency with what is in the parameter file.[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file[in] do_not_read: If present and true, do not read a value for this parameter, although it might be logged.[in] do_not_log: If present and true, do not log this parameter to the documentation files[in] timeunit: The number of seconds in a time unit for real-number input to be translated to a time.[in] layoutparam: If present and true, this parameter is logged in the layout parameter file[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file[in] log_as_date: If true, log the time_type in date format. The default is false.
-
subroutine mom_file_parser::get_param::get_param_int_array(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, fail_if_missing fail_if_missing, do_not_read do_not_read, do_not_log do_not_log, static_value static_value, layoutParam layoutParam, debuggingParam debuggingParam) This subroutine reads the values of an array of integer model parameters from a parameter file and logs them in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be reset from the parameter file[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] static_value: If this parameter is static, it takes this value, which can be compared for consistency with what is in the parameter file.[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file[in] do_not_read: If present and true, do not read a value for this parameter, although it might be logged.[in] do_not_log: If present and true, do not log this parameter to the documentation files[in] layoutparam: If present and true, this parameter is logged in the layout parameter file[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file
-
subroutine mom_file_parser::get_param::get_param_real_array(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, fail_if_missing fail_if_missing, do_not_read do_not_read, do_not_log do_not_log, debuggingParam debuggingParam, static_value static_value, scale scale, unscaled unscaled) This subroutine reads the values of an array of real model parameters from a parameter file and logs them in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file and logged[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] static_value: If this parameter is static, it takes this value, which can be compared for consistency with what is in the parameter file.[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file[in] do_not_read: If present and true, do not read a value for this parameter, although it might be logged.[in] do_not_log: If present and true, do not log this parameter to the documentation files[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file[in] scale: A scaling factor that the parameter is multiplied by before it is returned.[out] unscaled: The value of the parameter that would be returned without any multiplication by a scaling factor.
-
- type
Global positioning system (aka container for information to describe the grid)
Public Members
-
real
len_lon¶ The longitudinal or x-direction length of the domain.
-
real
len_lat¶ The latitudinal or y-direction length of the domain.
-
real
west_lon¶ The western longitude of the domain or the equivalent starting value for the x-axis.
-
real
south_lat¶ The southern latitude of the domain or the equivalent starting value for the y-axis.
-
real
rad_earth¶ The radius of the Earth [m].
-
real
lat_enhance_factor¶ The amount by which the meridional resolution is enhanced within LAT_EQ_ENHANCE of the equator.
-
real
lat_eq_enhance¶ The latitude range to the north and south of the equator over which the resolution is enhanced, in degrees.
-
logical
isotropic¶ If true, an isotropic grid on a sphere (also known as a Mercator grid) is used. With an isotropic grid, the meridional extent of the domain (LENLAT), the zonal extent (LENLON), and the number of grid points in each direction are not independent. In MOM the meridional extent is determined to fit the zonal extent and the number of grid points, while grid is perfectly isotropic.
-
logical
equator_reference¶ If true, the grid is defined to have the equator at the nearest q- or h- grid point to (-LOWLAT*NJGLOBAL/LENLAT).
-
integer
niglobal¶ The number of i-points in the global grid computational domain.
-
integer
njglobal¶ The number of j-points in the global grid computational domain.
-
real
-
interface
hchksum¶ Checksums an array (2d or 3d) staggered at tracer points.
Private Functions
-
subroutine
chksum_h_2d(array array, mesg mesg, HI HI, haloshift haloshift, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums a 2d array staggered at tracer points.
- Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] mesg: An identifying message[in] haloshift: The width of halos to check (default 0)[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
chksum_h_3d(array array, mesg mesg, HI HI, haloshift haloshift, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums a 3d array staggered at tracer points.
- Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] mesg: An identifying message[in] haloshift: The width of halos to check (default 0)[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
-
interface
hchksum_pair¶ Checksums a pair of arrays (2d or 3d) staggered at tracer points.
Private Functions
-
subroutine
chksum_pair_h_2d(mesg mesg, arrayA arrayA, arrayB arrayB, HI HI, haloshift haloshift, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums on a pair of 2d arrays staggered at tracer points.
- Parameters
[in] mesg: Identifying messages[in] hi: A horizontal index type[in] arraya: The first array to be checksummed[in] arrayb: The second array to be checksummed[in] haloshift: The width of halos to check (default 0)[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
chksum_pair_h_3d(mesg mesg, arrayA arrayA, arrayB arrayB, HI HI, haloshift haloshift, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums on a pair of 3d arrays staggered at tracer points.
- Parameters
[in] mesg: Identifying messages[in] hi: A horizontal index type[in] arraya: The first array to be checksummed[in] arrayb: The second array to be checksummed[in] haloshift: The width of halos to check (default 0)[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
- type
Container for horizontal index ranges for data, computational and global domains.
Public Members
-
integer
isc¶ The start i-index of cell centers within the computational domain.
-
integer
iec¶ The end i-index of cell centers within the computational domain.
-
integer
jsc¶ The start j-index of cell centers within the computational domain.
-
integer
jec¶ The end j-index of cell centers within the computational domain.
-
integer
isd¶ The start i-index of cell centers within the data domain.
-
integer
ied¶ The end i-index of cell centers within the data domain.
-
integer
jsd¶ The start j-index of cell centers within the data domain.
-
integer
jed¶ The end j-index of cell centers within the data domain.
-
integer
isg¶ The start i-index of cell centers within the global domain.
-
integer
ieg¶ The end i-index of cell centers within the global domain.
-
integer
jsg¶ The start j-index of cell centers within the global domain.
-
integer
jeg¶ The end j-index of cell centers within the global domain.
-
integer
iscb¶ The start i-index of cell vertices within the computational domain.
-
integer
iecb¶ The end i-index of cell vertices within the computational domain.
-
integer
jscb¶ The start j-index of cell vertices within the computational domain.
-
integer
jecb¶ The end j-index of cell vertices within the computational domain.
-
integer
isdb¶ The start i-index of cell vertices within the data domain.
-
integer
iedb¶ The end i-index of cell vertices within the data domain.
-
integer
jsdb¶ The start j-index of cell vertices within the data domain.
-
integer
jedb¶ The end j-index of cell vertices within the data domain.
-
integer
isgb¶ The start i-index of cell vertices within the global domain.
-
integer
iegb¶ The end i-index of cell vertices within the global domain.
-
integer
jsgb¶ The start j-index of cell vertices within the global domain.
-
integer
jegb¶ The end j-index of cell vertices within the global domain.
-
integer
idg_offset¶ The offset between the corresponding global and local i-indices.
-
integer
jdg_offset¶ The offset between the corresponding global and local j-indices.
-
logical
symmetric¶ True if symmetric memory is used.
-
integer
- type
Control structure for horizontal viscosity.
Unnamed Group
-
integer
id_diffu= -1¶ Diagnostic id.
-
integer
id_diffv= -1¶ Diagnostic id.
-
integer
id_ah_h= -1¶ Diagnostic id.
-
integer
id_ah_q= -1¶ Diagnostic id.
-
integer
id_kh_h= -1¶ Diagnostic id.
-
integer
id_kh_q= -1¶ Diagnostic id.
-
integer
id_gme_coeff_h= -1¶ Diagnostic id.
-
integer
id_gme_coeff_q= -1¶ Diagnostic id.
-
integer
id_vort_xy_q= -1¶ Diagnostic id.
-
integer
id_div_xx_h= -1¶ Diagnostic id.
-
integer
id_frictwork= -1¶ Diagnostic id.
-
integer
id_frictworkintz= -1¶ Diagnostic id.
-
integer
id_frictworkmax= -1¶ Diagnostic id.
-
integer
id_frictwork_diss= -1¶ Diagnostic id.
-
integer
id_frictwork_gme= -1¶ Diagnostic id.
Public Members
-
logical
laplacian¶ Use a Laplacian horizontal viscosity if true.
-
logical
biharmonic¶ Use a biharmonic horizontal viscosity if true.
-
logical
no_slip¶ If true, no slip boundary conditions are used. Otherwise free slip boundary conditions are assumed. The implementation of the free slip boundary conditions on a C-grid is much cleaner than the no slip boundary conditions. The use of free slip b.c.s is strongly encouraged. The no slip b.c.s are not implemented with the biharmonic viscosity.
-
logical
bound_kh¶ If true, the Laplacian coefficient is locally limited to guarantee stability.
-
logical
better_bound_kh¶ If true, use a more careful bounding of the Laplacian viscosity to guarantee stability.
-
logical
bound_ah¶ If true, the biharmonic coefficient is locally limited to guarantee stability.
-
logical
better_bound_ah¶ If true, use a more careful bounding of the biharmonic viscosity to guarantee stability.
-
real
bound_coef¶ The nondimensional coefficient of the ratio of the viscosity bounds to the theoretical maximum for stability without considering other terms [nondim]. The default is 0.8.
-
logical
smagorinsky_kh¶ If true, use Smagorinsky nonlinear eddy viscosity. KH is the background value.
-
logical
smagorinsky_ah¶ If true, use a biharmonic form of Smagorinsky nonlinear eddy viscosity. AH is the background.
-
logical
leith_kh¶ If true, use 2D Leith nonlinear eddy viscosity. KH is the background value.
-
logical
modified_leith¶ If true, use extra component of Leith viscosity to damp divergent flow. To use, still set Leith_Kh=.TRUE.
-
logical
use_beta_in_leith¶ If true, includes the beta term in the Leith viscosity.
-
logical
leith_ah¶ If true, use a biharmonic form of 2D Leith nonlinear eddy viscosity. AH is the background.
-
logical
use_qg_leith_visc¶ If true, use QG Leith nonlinear eddy viscosity. KH is the background value.
-
logical
bound_coriolis¶ If true & SMAGORINSKY_AH is used, the biharmonic viscosity is modified to include a term that scales quadratically with the velocity shears.
-
logical
use_kh_bg_2d¶ Read 2d background viscosity from a file.
-
real
kh_bg_min¶ The minimum value allowed for Laplacian horizontal viscosity [m2 T-1 ~> m2 s-1]. The default is 0.0.
-
logical
use_land_mask¶ Use the land mask for the computation of thicknesses at velocity locations. This eliminates the dependence on arbitrary values over land or outside of the domain. Default is False to maintain answers with legacy experiments but should be changed to True for new experiments.
-
logical
anisotropic¶ If true, allow anisotropic component to the viscosity.
-
real
kh_aniso¶ The anisotropic viscosity [m2 T-1 ~> m2 s-1].
-
logical
dynamic_aniso¶ If true, the anisotropic viscosity is recomputed as a function of state. This is set depending on ANISOTROPIC_MODE.
-
logical
res_scale_meke¶ If true, the viscosity contribution from MEKE is scaled by the resolution function.
-
logical
use_gme¶ If true, use GME backscatter scheme.
-
logical
answers_2018¶ If true, use the order of arithmetic and expressions that recover the answers from the end of 2018. Otherwise, use updated and more robust forms of the same expressions.
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::kh_bg_xx The background Laplacian viscosity at h points [m2 T-1 ~> m2 s-1]. The actual viscosity may be the larger of this viscosity and the Smagorinsky and Leith viscosities.
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::kh_bg_2d The background Laplacian viscosity at h points [m2 T-1 ~> m2 s-1]. The actual viscosity may be the larger of this viscosity and the Smagorinsky and Leith viscosities.
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::ah_bg_xx The background biharmonic viscosity at h points [m4 T-1 ~> m4 s-1]. The actual viscosity may be the larger of this viscosity and the Smagorinsky and Leith viscosities.
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::biharm5_const2_xx A constant relating the biharmonic viscosity to the square of the velocity shear [m4 s]. This value is set to be the magnitude of the Coriolis terms once the velocity differences reach a value of order 1/2 MAXVEL.
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::reduction_xx The amount by which stresses through h points are reduced due to partial barriers. Nondimensional.
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::kh_max_xx The maximum permitted Laplacian viscosity [m2 T-1 ~> m2 s-1].
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::ah_max_xx The maximum permitted biharmonic viscosity [m4 T-1 ~> m4 s-1].
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::n1n2_h Factor n1*n2 in the anisotropic direction tensor at h-points.
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::n1n1_m_n2n2_h Factor n1**2-n2**2 in the anisotropic direction tensor at h-points.
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::kh_bg_xy The background Laplacian viscosity at q points [m2 T-1 ~> m2 s-1]. The actual viscosity may be the larger of this viscosity and the Smagorinsky and Leith viscosities.
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::ah_bg_xy The background biharmonic viscosity at q points [m4 T-1 ~> m4 s-1]. The actual viscosity may be the larger of this viscosity and the Smagorinsky and Leith viscosities.
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::biharm5_const2_xy A constant relating the biharmonic viscosity to the square of the velocity shear [m4 s]. This value is set to be the magnitude of the Coriolis terms once the velocity differences reach a value of order 1/2 MAXVEL.
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::reduction_xy The amount by which stresses through q points are reduced due to partial barriers [nondim].
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::kh_max_xy The maximum permitted Laplacian viscosity [m2 T-1 ~> m2 s-1].
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::ah_max_xy The maximum permitted biharmonic viscosity [m4 T-1 ~> m4 s-1].
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::n1n2_q Factor n1*n2 in the anisotropic direction tensor at q-points.
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::n1n1_m_n2n2_q Factor n1**2-n2**2 in the anisotropic direction tensor at q-points.
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::dx2h Pre-calculated dx^2 at h points [m2].
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::dy2h Pre-calculated dy^2 at h points [m2].
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::dx_dyt Pre-calculated dx/dy at h points [nondim].
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::dy_dxt Pre-calculated dy/dx at h points [nondim].
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::dx2q Pre-calculated dx^2 at q points [m2].
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::dy2q Pre-calculated dy^2 at q points [m2].
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::dx_dybu Pre-calculated dx/dy at q points [nondim].
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::dy_dxbu Pre-calculated dy/dx at q points [nondim].
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::idx2dycu 1/(dx^2 dy) at u points [m-3]
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::idxdy2u 1/(dx dy^2) at u points [m-3]
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::idx2dycv 1/(dx^2 dy) at v points [m-3]
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::idxdy2v 1/(dx dy^2) at v points [m-3]
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::laplac2_const_xx Laplacian metric-dependent constants [m2].
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::biharm5_const_xx Biharmonic metric-dependent constants [m5].
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::laplac3_const_xx Laplacian metric-dependent constants [m3].
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::biharm_const_xx Biharmonic metric-dependent constants [m4].
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::biharm_const2_xx Biharmonic metric-dependent constants [T m4 ~> s m4].
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::laplac2_const_xy Laplacian metric-dependent constants [m2].
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::biharm5_const_xy Biharmonic metric-dependent constants [m5].
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::laplac3_const_xy Laplacian metric-dependent constants [m3].
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::biharm_const_xy Biharmonic metric-dependent constants [m4].
-
real, dimension( : , : ), allocatable mom_hor_visc::hor_visc_cs::biharm_const2_xy Biharmonic metric-dependent constants [T m4 ~> s m4].
-
type(diag_ctrl), pointer mom_hor_visc::hor_visc_cs::diag => NULL() structure to regulate diagnostics
-
integer
-
interface
horiz_interp_and_extrap_tracer¶ Extrapolate and interpolate data.
Private Functions
-
subroutine
horiz_interp_and_extrap_tracer_record(filename filename, varnam varnam, conversion conversion, recnum recnum, G G, tr_z tr_z, mask_z mask_z, z_in z_in, z_edges_in z_edges_in, missing_value missing_value, reentrant_x reentrant_x, tripolar_n tripolar_n, homogenize homogenize, m_to_Z m_to_Z)¶ Extrapolate and interpolate from a file record.
- Parameters
[in] filename: Path to file containing tracer to be interpolated.[in] varnam: Name of tracer in filee.[in] conversion: Conversion factor for tracer.[in] recnum: Record number of tracer to be read.[inout] g: Grid objecttr_z: pointer to allocatable tracer array on local model grid and input-file vertical levels.mask_z: pointer to allocatable tracer mask array on local model grid and input-file vertical levels.z_in: Cell grid values for input data.z_edges_in: Cell grid edge values for input data.[out] missing_value: The missing value in the returned array.[in] reentrant_x: If true, this grid is reentrant in the x-direction[in] tripolar_n: If true, this is a northern tripolar grid[in] homogenize: If present and true, horizontally homogenize data to produce perfectly “flat” initial conditions[in] m_to_z: A conversion factor from meters to the units of depth. If missing, GbathyT must be in m.
-
subroutine
horiz_interp_and_extrap_tracer_fms_id(fms_id fms_id, Time Time, conversion conversion, G G, tr_z tr_z, mask_z mask_z, z_in z_in, z_edges_in z_edges_in, missing_value missing_value, reentrant_x reentrant_x, tripolar_n tripolar_n, homogenize homogenize, m_to_Z m_to_Z)¶ Extrapolate and interpolate using a FMS time interpolation handle.
- Parameters
[in] fms_id: A unique id used by the FMS time interpolator[in] time: A FMS time type[in] conversion: Conversion factor for tracer.[inout] g: Grid objecttr_z: pointer to allocatable tracer array on local model grid and native vertical levels.mask_z: pointer to allocatable tracer mask array on local model grid and native vertical levels.z_in: Cell grid values for input data.z_edges_in: Cell grid edge values for input data. (Intent out)[out] missing_value: The missing value in the returned array.[in] reentrant_x: If true, this grid is reentrant in the x-direction[in] tripolar_n: If true, this is a northern tripolar grid[in] homogenize: If present and true, horizontally homogenize data to produce perfectly “flat” initial conditions[in] m_to_z: A conversion factor from meters to the units of depth. If missing, GbathyT must be in m.
-
subroutine
- type
Control structure containing required parameters for the HyCOM coordinate.
Public Members
-
integer
nk¶ Number of layers/levels in generated grid.
-
real, dimension(:), allocatable coord_hycom::hycom_cs::coordinateresolution Nominal near-surface resolution.
-
real, dimension(:), allocatable coord_hycom::hycom_cs::target_density Nominal density of interfaces.
-
real, dimension(:), allocatable coord_hycom::hycom_cs::max_interface_depths Maximum depths of interfaces.
-
real, dimension(:), allocatable coord_hycom::hycom_cs::max_layer_thickness Maximum thicknesses of layers.
-
type(interp_cs_type) coord_hycom::hycom_cs::interp_cs Interpolation control structure.
-
integer
- type
Control structure that contains ice shelf parameters and diagnostics handles.
Unnamed Group
-
integer
id_melt= -1¶ Diagnostic handles.
-
integer
id_exch_vel_s= -1¶ Diagnostic handles.
-
integer
id_exch_vel_t= -1¶ Diagnostic handles.
-
integer
id_tfreeze= -1¶ Diagnostic handles.
-
integer
id_tfl_shelf= -1¶ Diagnostic handles.
-
integer
id_thermal_driving= -1¶ Diagnostic handles.
-
integer
id_haline_driving= -1¶ Diagnostic handles.
-
integer
id_u_ml= -1¶ Diagnostic handles.
-
integer
id_v_ml= -1¶ Diagnostic handles.
-
integer
id_sbdry= -1¶ Diagnostic handles.
-
integer
id_h_shelf= -1¶ Diagnostic handles.
-
integer
id_h_mask= -1¶ Diagnostic handles.
-
integer
id_surf_elev= -1¶ Diagnostic handles.
-
integer
id_bathym= -1¶ Diagnostic handles.
-
integer
id_area_shelf_h= -1¶ Diagnostic handles.
-
integer
id_ustar_shelf= -1¶ Diagnostic handles.
-
integer
id_shelf_mass= -1¶ Diagnostic handles.
-
integer
id_mass_flux= -1¶ Diagnostic handles.
Public Members
-
type(mom_restart_cs), pointer mom_ice_shelf::ice_shelf_cs::restart_csp => NULL() A pointer to the restart control structure for the ice shelves.
-
type(ocean_grid_type) mom_ice_shelf::ice_shelf_cs::grid Grid for the ice-shelf model.
-
type(unit_scale_type), pointer mom_ice_shelf::ice_shelf_cs::us => NULL() A structure containing various unit conversion factors.
-
type(ocean_grid_type), pointer mom_ice_shelf::ice_shelf_cs::ocn_grid => NULL() A pointer to the ocean model grid The rest is private.
-
real
flux_factor= 1.0¶ A factor that can be used to turn off ice shelf melting (flux_factor = 0) [nondim].
-
character(len=128) mom_ice_shelf::ice_shelf_cs::restart_output_dir = ' ' The directory in which to write restart files.
-
type(ice_shelf_state), pointer mom_ice_shelf::ice_shelf_cs::iss => NULL() A structure with elements that describe the ice-shelf state.
-
type(ice_shelf_dyn_cs), pointer mom_ice_shelf::ice_shelf_cs::dcs => NULL() The control structure for the ice-shelf dynamics.
-
real, dimension(:,:), pointer mom_ice_shelf::ice_shelf_cs::utide => NULL() tidal velocity [m s-1]
-
real
ustar_bg¶ A minimum value for ustar under ice shelves [Z T-1 ~> m s-1].
-
real
cdrag¶ drag coefficient under ice shelves [nondim].
-
real
g_earth¶ The gravitational acceleration [m s-2].
-
real
cp¶ The heat capacity of sea water [J kg-1 degC-1].
-
real
rho0¶ A reference ocean density [kg m-3].
-
real
cp_ice¶ The heat capacity of fresh ice [J kg-1 degC-1].
-
real
gamma_t¶ The (fixed) turbulent exchange velocity in the 2-equation formulation [m s-1].
-
real
salin_ice¶ The salinity of shelf ice [ppt].
-
real
temp_ice¶ The core temperature of shelf ice [degC].
-
real
kv_ice¶ The viscosity of ice [m2 s-1].
-
real
density_ice¶ A typical density of ice [kg m-3].
-
real
rho_ice¶ Nominal ice density [kg m-2 Z-1 ~> kg m-3].
-
real
kv_molec¶ The molecular kinematic viscosity of sea water [m2 s-1].
-
real
kd_molec_salt¶ The molecular diffusivity of salt [m2 s-1].
-
real
kd_molec_temp¶ The molecular diffusivity of heat [m2 s-1].
-
real
lat_fusion¶ The latent heat of fusion [J kg-1].
-
real
gamma_t_3eq¶ Nondimensional heat-transfer coefficient, used in the 3Eq. formulation This number should be specified by the user.
-
real
col_thick_melt_threshold¶ if the mixed layer is below this threshold, melt rate
-
logical
mass_from_file¶ Read the ice shelf mass from a file every dt.
-
real
time_step¶ this is the shortest timestep that the ice shelf sees, and is equal to the forcing timestep (it is passed in when the shelf is initialized - so need to reorganize MOM driver. it will be the prognistic timestep … maybe.
-
logical
solo_ice_sheet¶ whether the ice model is running without being coupled to the ocean
-
logical
gl_regularize¶ whether to regularize the floatation condition at the grounding line a la Goldberg Holland Schoof 2009
-
logical
gl_couple¶ whether to let the floatation condition be determined by ocean column thickness means update_OD_ffrac will be called (note: GL_regularize and GL_couple should be exclusive)
-
real
density_ocean_avg¶ this does not affect ocean circulation OR thermodynamics it is to estimate the gravitational driving force at the shelf front (until we think of a better way to do it, but any difference will be negligible)
-
logical
calve_to_mask¶ If true, calve any ice that passes outside of a masked area.
-
real
min_thickness_simple_calve¶ min. ice shelf thickness criteria for calving [Z ~> m].
-
real
t0¶ temperature at ocean surface in the restoring region [degC]
-
real
s0¶ Salinity at ocean surface in the restoring region [ppt].
-
real
input_flux¶ Ice volume flux at an upstream open boundary [m3 s-1].
-
real
input_thickness¶ Ice thickness at an upstream open boundary [m].
-
type(time_type) mom_ice_shelf::ice_shelf_cs::time The component’s time.
-
type(eos_type), pointer mom_ice_shelf::ice_shelf_cs::eqn_of_state => NULL() Type that indicates the equation of state to use.
-
logical
active_shelf_dynamics¶ True if the ice shelf mass changes as a result the dynamic ice-shelf model.
-
logical
override_shelf_movement¶ If true, user code specifies the shelf movement instead of using the dynamic ice-shelf mode.
-
logical
isthermo¶ True if the ice shelf can exchange heat and mass with the underlying ocean.
-
logical
threeeq¶ If true, the 3 equation consistency equations are used to calculate the flux at the ocean-ice interface.
-
logical
insulator¶ If true, ice shelf is a perfect insulator.
-
logical
const_gamma¶ If true, gamma_T is specified by the user.
-
logical
find_salt_root¶ If true, if true find Sbdry using a quadratic eq.
-
logical
constant_sea_level¶ if true, apply an evaporative, heat and salt fluxes. It will avoid large increase in sea level.
-
real
cutoff_depth¶ depth above which melt is set to zero (>= 0).
-
real
lambda1¶ liquidus coeff., Needed if find_salt_root = true
-
real
lambda2¶ liquidus coeff., Needed if find_salt_root = true
-
real
lambda3¶ liquidus coeff., Needed if find_salt_root = true
-
integer
id_read_mass¶ An integer handle used in time interpolation of the ice shelf mass read from a file.
-
integer
id_read_area¶ An integer handle used in time interpolation of the ice shelf mass read from a file.
-
type(diag_ctrl), pointer mom_ice_shelf::ice_shelf_cs::diag => NULL() A structure that is used to control diagnostic output.
-
type(user_ice_shelf_cs), pointer mom_ice_shelf::ice_shelf_cs::user_cs => NULL() A pointer to the control structure for user-supplied modifications to the ice shelf code.
-
logical
debug¶ If true, write verbose checksums for debugging purposes and use reproducible sums.
-
integer
- type
The control structure for the ice shelf dynamics.
Unnamed Group
-
integer
id_u_shelf= -1¶ Diagnostic handles.
-
integer
id_v_shelf= -1¶ Diagnostic handles.
-
integer
id_t_shelf= -1¶ Diagnostic handles.
-
integer
id_float_frac= -1¶ Diagnostic handles.
-
integer
id_col_thick= -1¶ Diagnostic handles.
-
integer
id_od_av= -1¶ Diagnostic handles.
-
integer
id_u_mask= -1¶ Diagnostic handles.
-
integer
id_v_mask= -1¶ Diagnostic handles.
-
integer
id_t_mask= -1¶ Diagnostic handles.
-
type(diag_ctrl), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::diag => NULL() A structure that is used to control diagnostic output.
Public Members
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::u_shelf => NULL() the zonal (?) velocity of the ice shelf/sheet on q-points (B grid) [m s-1]??
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::v_shelf => NULL() the meridional velocity of the ice shelf/sheet on q-points (B grid) [m s-1]??
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::u_face_mask => NULL() mask for velocity boundary conditions on the C-grid u-face - this is because the FEM cares about FACES THAT GET INTEGRATED OVER, not vertices. Will represent boundary conditions on computational boundary (or permanent boundary between fast-moving and near-stagnant ice FOR NOW: 1=interior bdry, 0=no-flow boundary, 2=stress bdry condition, 3=inhomogeneous dirichlet boundary, 4=flux boundary: at these faces a flux will be specified which will override velocities; a homogeneous velocity condition will be specified (this seems to give the solver less difficulty)
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::v_face_mask => NULL() A mask for velocity boundary conditions on the C-grid v-face, with valued defined similarly to u_face_mask.
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::u_face_mask_bdry => NULL() A duplicate copy of u_face_mask?
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::v_face_mask_bdry => NULL() A duplicate copy of v_face_mask?
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::u_flux_bdry_val => NULL() The ice volume flux into the cell through open boundary u-faces (where u_face_mask=4) [Z m2 s-1 ~> m3 s-1]??
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::v_flux_bdry_val => NULL() The ice volume flux into the cell through open boundary v-faces (where v_face_mask=4) [Z m2 s-1 ~> m3 s-1]??
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::umask => NULL() u-mask on the actual degrees of freedom (B grid) 1=normal node, 3=inhomogeneous boundary node, 0 - no flow node (will also get ice-free nodes)
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::vmask => NULL() v-mask on the actual degrees of freedom (B grid) 1=normal node, 3=inhomogeneous boundary node, 0 - no flow node (will also get ice-free nodes)
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::calve_mask => NULL() a mask to prevent the ice shelf front from advancing past its initial position (but it may retreat)
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::t_shelf => NULL() Veritcally integrated temperature in the ice shelf/stream, on corner-points (B grid) [degC].
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::tmask => NULL() A mask on tracer points that is 1 where there is ice.
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::ice_visc => NULL() Glen’s law ice viscosity, perhaps in [m].
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::thickness_bdry_val => NULL() The ice thickness at an inflowing boundary [Z ~> m].
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::u_bdry_val => NULL() The zonal ice velocity at inflowing boundaries [m s-1]??
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::v_bdry_val => NULL() The meridional ice velocity at inflowing boundaries [m s-1]??
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::h_bdry_val => NULL() The ice thickness at inflowing boundaries [m].
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::t_bdry_val => NULL() The ice temperature at inflowing boundaries [degC].
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::taub_beta_eff => NULL() nonlinear part of “linearized” basal stress. The exact form depends on basal law exponent and/or whether flow is “hybridized” a la Goldberg 2011
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::od_rt => NULL() A running total for calculating OD_av.
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::float_frac_rt => NULL() A running total for calculating float_frac.
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::od_av => NULL() The time average open ocean depth [Z ~> m].
-
real, dimension(:,:), pointer mom_ice_shelf_dynamics::ice_shelf_dyn_cs::float_frac => NULL() Fraction of the time a cell is “exposed”, i.e. the column thickness is below a threshold.
-
integer
od_rt_counter= 0¶ A counter of the number of contributions to OD_rt.
-
real
velocity_update_time_step¶ The time interval over which to update the ice shelf velocity using the nonlinear elliptic equation, or 0 to update every timestep [s].
-
real
elapsed_velocity_time¶ The elapsed time since the ice velocies were last udated [s].
-
real
g_earth¶ The gravitational acceleration [m s-2].
-
real
density_ice¶ A typical density of ice [kg m-3].
-
logical
gl_regularize¶ Specifies whether to regularize the floatation condition at the grounding line as in Goldberg Holland Schoof 2009.
-
integer
n_sub_regularize¶ partition of cell over which to integrate for interpolated grounding line the (rectangular) is divided into nxn equally-sized rectangles, over which basal contribution is integrated (iterative quadrature)
-
logical
gl_couple¶ whether to let the floatation condition be determined by ocean column thickness means update_OD_ffrac will be called (note: GL_regularize and GL_couple should be exclusive)
-
real
cfl_factor¶ A factor used to limit subcycled advective timestep in uncoupled runs i.e. dt <= CFL_factor * min(dx / u)
-
real
a_glen_isothermal¶ Ice viscosity parameter in Glen’s Lawa, [Pa-1/3 year].
-
real
n_glen¶ Nonlinearity exponent in Glen’s Law.
-
real
eps_glen_min¶ Min. strain rate to avoid infinite Glen’s law viscosity, [year-1].
-
real
c_basal_friction¶ Ceofficient in sliding law tau_b = C u^(n_basal_friction), in units=”Pa (m-a)-(n_basal_friction)
-
real
n_basal_friction¶ Exponent in sliding law tau_b = C u^(m_slide)
-
real
density_ocean_avg¶ This does not affect ocean circulation or thermodynamics. It is used to estimate the gravitational driving force at the shelf front (until we think of a better way to do it, but any difference will be negligible).
-
real
thresh_float_col_depth¶ The water column depth over which the shelf if considered to be floating.
-
logical
moving_shelf_front¶ Specify whether to advance shelf front (and calve).
-
logical
calve_to_mask¶ If true, calve off the ice shelf when it passes the edge of a mask.
-
real
min_thickness_simple_calve¶ min. ice shelf thickness criteria for calving [Z ~> m].
-
real
cg_tolerance¶ The tolerance in the CG solver, relative to initial residual, that deterimnes when to stop the conguage gradient iterations.
-
real
nonlinear_tolerance¶ The fractional nonlinear tolerance, relative to the initial error, that sets when to stop the iterative velocity solver.
-
integer
cg_max_iterations¶ The maximum number of iterations that can be used in the CG solver.
-
integer
nonlin_solve_err_mode¶ 1: exit vel solve based on nonlin residual 2: exit based on “fixed point” metric (|u - u_last| / |u| < tol where | | is infty-norm
-
logical
use_reproducing_sums¶ Use reproducing global sums.
-
logical
debug¶ If true, write verbose checksums for debugging purposes and use reproducible sums.
-
logical
module_is_initialized= .false.¶ True if this module has been initialized.
-
integer
- type
Structure that describes the ice shelf state.
Public Members
-
real, dimension(:,:), pointer mom_ice_shelf_state::ice_shelf_state::mass_shelf => NULL() The mass per unit area of the ice shelf or sheet [kg m-2].
-
real, dimension(:,:), pointer mom_ice_shelf_state::ice_shelf_state::area_shelf_h => NULL() The area per cell covered by the ice shelf [m2].
-
real, dimension(:,:), pointer mom_ice_shelf_state::ice_shelf_state::h_shelf => NULL() the thickness of the shelf [m], redundant with mass but may
-
real, dimension(:,:), pointer mom_ice_shelf_state::ice_shelf_state::hmask => NULL() Mask used to indicate ice-covered or partiall-covered cells.
-
real, dimension(:,:), pointer mom_ice_shelf_state::ice_shelf_state::tflux_ocn => NULL() The UPWARD sensible ocean heat flux at the.
-
real, dimension(:,:), pointer mom_ice_shelf_state::ice_shelf_state::salt_flux => NULL() The downward salt flux at the ocean-ice.
-
real, dimension(:,:), pointer mom_ice_shelf_state::ice_shelf_state::water_flux => NULL() The net downward liquid water flux at the.
-
real, dimension(:,:), pointer mom_ice_shelf_state::ice_shelf_state::tflux_shelf => NULL() The UPWARD diffusive heat flux in the ice.
-
real, dimension(:,:), pointer mom_ice_shelf_state::ice_shelf_state::tfreeze => NULL() The freezing point potential temperature.
-
- type
The control structure for the ideal_age_tracer package.
Public Members
-
integer
ntr¶ The number of tracers that are actually used.
-
logical
coupled_tracers= .false.¶ These tracers are not offered to the coupler.
-
integer
nkml¶ The number of layers in the mixed layer. The ideal.
-
character(len=200) ideal_age_example::ideal_age_tracer_cs::ic_file The file in which the age-tracer initial values can be found, or an empty string for internal initialization.
-
logical
z_ic_file¶ If true, the IC_file is in Z-space. The default is false.
-
type(time_type), pointer ideal_age_example::ideal_age_tracer_cs::time => NULL() A pointer to the ocean model’s clock.
-
type(tracer_registry_type), pointer ideal_age_example::ideal_age_tracer_cs::tr_reg => NULL() A pointer to the tracer registry.
-
real, dimension(:,:,:,:), pointer ideal_age_example::ideal_age_tracer_cs::tr => NULL() The array of tracers used in this package, in g m-3?
-
real, dimension(ntr_max) ideal_age_example::ideal_age_tracer_cs::ic_val = 0.0 The (uniform) initial condition value.
-
real, dimension(ntr_max) ideal_age_example::ideal_age_tracer_cs::young_val = 0.0 The value assigned to tr at the surface.
-
real, dimension(ntr_max) ideal_age_example::ideal_age_tracer_cs::land_val = -1.0 The value of tr used where land is masked out.
-
real, dimension(ntr_max) ideal_age_example::ideal_age_tracer_cs::sfc_growth_rate The exponential growth rate for the surface value [year-1].
-
real, dimension(ntr_max) ideal_age_example::ideal_age_tracer_cs::tracer_start_year The year in which tracers start aging, or at which the surface value equals young_val, in years.
-
logical
tracers_may_reinit¶ If true, these tracers be set up via the initialization code if they are not found in the restart files.
-
logical, dimension(ntr_max) ideal_age_example::ideal_age_tracer_cs::tracer_ages Indicates whether each tracer ages.
-
integer, dimension(ntr_max) ideal_age_example::ideal_age_tracer_cs::ind_tr Indices returned by aof_set_coupler_flux if it is used and the surface tracer concentrations are to be provided to the coupler.
-
type(diag_ctrl), pointer ideal_age_example::ideal_age_tracer_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
type(mom_restart_cs), pointer ideal_age_example::ideal_age_tracer_cs::restart_csp => NULL() A pointer to the restart controls structure.
-
type(vardesc), dimension(ntr_max) ideal_age_example::ideal_age_tracer_cs::tr_desc Descriptions and metadata for the tracers.
-
integer
- type
Container for parameters describing idealized wind structure.
Public Members
-
real
rho_a¶ Mean air density [kg m-3].
-
real
pressure_ambient¶ Pressure at surface of ambient air [Pa].
-
real
pressure_central¶ Pressure at surface at hurricane center [Pa].
-
real
rad_max_wind¶ Radius of maximum winds [m].
-
real
max_windspeed¶ Maximum wind speeds [m s-1].
-
real
hurr_translation_spd¶ Hurricane translation speed [m s-1].
-
real
hurr_translation_dir¶ Hurricane translation speed [m s-1].
-
real
gustiness¶ Gustiness (optional, used in u*) [m s-1].
-
real
rho0¶ A reference ocean density [kg m-3].
-
real
hurr_cen_y0¶ The initial y position of the hurricane This experiment is conducted in a Cartesian grid and this is assumed to be in meters [m].
-
real
hurr_cen_x0¶ The initial x position of the hurricane This experiment is conducted in a Cartesian grid and this is assumed to be in meters [m].
-
real
holland_a¶ Parameter ‘A’ from the Holland formula.
-
real
holland_b¶ Parameter ‘B’ from the Holland formula.
-
real
holland_axbxdp¶ ‘A’ x ‘B’ x (Pressure Ambient-Pressure central) for the Holland prorfile calculation
-
logical
relative_tau¶ A logical to take difference between wind and surface currents to compute the stress.
-
logical
scm_mode¶ If true this being used in Single Column Model mode.
-
logical
br_bench¶ A “benchmark” configuration (which is meant to provide identical wind to reproduce a previous experiment, where that wind formula contained an error)
-
real
dy_from_center¶ (Fixed) distance in y from storm center path [m]
-
real
pi¶ Mathematical constant.
-
real
deg2rad¶ Mathematical constant.
-
real
-
interface
initialize_ale_sponge¶ Ddetermine the number of points which are within sponges in this computational domain.
Only points that have positive values of Iresttime and which mask2dT indicates are ocean points are included in the sponges. It also stores the target interface heights.
Private Functions
-
subroutine
initialize_ale_sponge_fixed(Iresttime Iresttime, G G, param_file param_file, CS CS, data_h data_h, nz_data nz_data)¶ This subroutine determines the number of points which are within sponges in this computational domain. Only points that have positive values of Iresttime and which mask2dT indicates are ocean points are included in the sponges. It also stores the target interface heights.
- Parameters
[in] g: The ocean’s grid structure.[in] nz_data: The total number of sponge input layers.[in] iresttime: The inverse of the restoring time [s-1].[in] param_file: A structure indicating the open file to parse for model parameter values.cs: A pointer that is set to point to the control structure for this module (in/out).[in] data_h: The thicknesses of the sponge input layers [H ~> m or kg m-2].
-
subroutine
initialize_ale_sponge_varying(Iresttime Iresttime, G G, param_file param_file, CS CS)¶ This subroutine determines the number of points which are within sponges in this computational domain. Only points that have positive values of Iresttime and which mask2dT indicates are ocean points are included in the sponges.
- Parameters
[in] g: The ocean’s grid structure.[in] iresttime: The inverse of the restoring time [s-1].[in] param_file: A structure indicating the open file to parse for model parameter values.cs: A pointer that is set to point to the control structure for this module (in/out).
-
subroutine
- type
This control structure has parameters for the MOM_internal_tides module.
Unnamed Group
-
integer
id_tot_en= -1¶ Diag handles.
-
integer
id_tke_itidal_input= -1¶ Diag handles.
-
integer
id_itide_drag= -1¶ Diag handles.
-
integer
id_refl_pref= -1¶ Diag handles.
-
integer
id_refl_ang= -1¶ Diag handles.
-
integer
id_land_mask= -1¶ Diag handles.
-
integer
id_dx_cv= -1¶ Diag handles.
-
integer
id_dy_cu= -1¶ Diag handles.
-
integer
id_tot_leak_loss= -1¶ Diag handles.
-
integer
id_tot_quad_loss= -1¶ Diag handles.
-
integer
id_tot_itidal_loss= -1¶ Diag handles.
-
integer
id_tot_froude_loss= -1¶ Diag handles.
-
integer
id_tot_allprocesses_loss= -1¶ Diag handles.
-
integer, dimension(:,:), allocatable mom_internal_tides::int_tide_cs::id_en_mode Diag handles.
-
integer, dimension(:,:), allocatable mom_internal_tides::int_tide_cs::id_itidal_loss_mode Diag handles.
-
integer, dimension(:,:), allocatable mom_internal_tides::int_tide_cs::id_allprocesses_loss_mode Diag handles.
-
integer, dimension(:,:), allocatable mom_internal_tides::int_tide_cs::id_ub_mode Diag handles.
-
integer, dimension(:,:), allocatable mom_internal_tides::int_tide_cs::id_cp_mode Diag handles.
-
integer, dimension(:,:), allocatable mom_internal_tides::int_tide_cs::id_en_ang_mode Diag handles.
-
integer, dimension(:,:), allocatable mom_internal_tides::int_tide_cs::id_itidal_loss_ang_mode Diag handles.
Public Members
-
logical
do_int_tides¶ If true, use the internal tide code.
-
integer
nfreq= 0¶ The number of internal tide frequency bands.
-
integer
nmode= 1¶ The number of internal tide vertical modes.
-
integer
nangle= 24¶ The number of internal tide angular orientations.
-
integer
energized_angle= -1¶ If positive, only this angular band is energized for debugging purposes.
-
logical
corner_adv¶ If true, use a corner advection rather than PPM.
-
logical
upwind_1st¶ If true, use a first-order upwind scheme.
-
logical
simple_2nd¶ If true, use a simple second order (arithmetic mean) interpolation of the edge values instead of the higher order interpolation.
-
logical
vol_cfl¶ If true, use the ratio of the open face lengths to the tracer cell areas when estimating CFL numbers. Without aggress_adjust, the default is false; it is always true with aggress_adjust.
-
logical
use_ppmang¶ If true, use PPM for advection of energy in angular space.
-
real, dimension(:,:), allocatable mom_internal_tides::int_tide_cs::refl_angle local coastline/ridge/shelf angles read from file
-
real
nullangle= -999.9¶ placeholder value in cells with no reflection
-
real, dimension(:,:), allocatable mom_internal_tides::int_tide_cs::refl_pref partial reflection coeff for each “coast cell”
-
logical, dimension(:,:), allocatable mom_internal_tides::int_tide_cs::refl_pref_logical true if reflecting cell with partial reflection
-
logical, dimension(:,:), allocatable mom_internal_tides::int_tide_cs::refl_dbl identifies reflection cells where double reflection is possible (i.e. ridge cells)
-
real, dimension(:,:,:,:), allocatable mom_internal_tides::int_tide_cs::cp horizontal phase speed [m s-1]
-
real, dimension(:,:,:,:,:), allocatable mom_internal_tides::int_tide_cs::tke_leak_loss energy lost due to misc background processes [W m-2]
-
real, dimension(:,:,:,:,:), allocatable mom_internal_tides::int_tide_cs::tke_quad_loss energy lost due to quadratic bottom drag [W m-2]
-
real, dimension(:,:,:,:,:), allocatable mom_internal_tides::int_tide_cs::tke_froude_loss energy lost due to wave breaking [W m-2]
-
real, dimension(:,:), allocatable mom_internal_tides::int_tide_cs::tke_itidal_loss_fixed fixed part of the energy lost due to small-scale drag [kg Z-2 ~> kg m-2] here; will be multiplied by N and En to get into [W m-2]
-
real, dimension(:,:,:,:,:), allocatable mom_internal_tides::int_tide_cs::tke_itidal_loss energy lost due to small-scale wave drag [W m-2]
-
real, dimension(:,:), allocatable mom_internal_tides::int_tide_cs::tot_leak_loss Energy loss rates due to misc bakground processes, summed over angle, frequency and mode [W m-2].
-
real, dimension(:,:), allocatable mom_internal_tides::int_tide_cs::tot_quad_loss Energy loss rates due to quadratic bottom drag, summed over angle, frequency and mode [W m-2].
-
real, dimension(:,:), allocatable mom_internal_tides::int_tide_cs::tot_itidal_loss Energy loss rates due to small-scale drag, summed over angle, frequency and mode [W m-2].
-
real, dimension(:,:), allocatable mom_internal_tides::int_tide_cs::tot_froude_loss Energy loss rates due to wave breaking, summed over angle, frequency and mode [W m-2].
-
real, dimension(:,:), allocatable mom_internal_tides::int_tide_cs::tot_allprocesses_loss Energy loss rates due to all processes, summed over angle, frequency and mode [W m-2].
-
real
q_itides¶ fraction of local dissipation [nondim]
-
real
en_sum¶ global sum of energy for use in debugging
-
type(time_type), pointer mom_internal_tides::int_tide_cs::time => NULL() A pointer to the model’s clock.
-
character(len=200) mom_internal_tides::int_tide_cs::inputdir directory to look for coastline angle file
-
real
decay_rate¶ A constant rate at which internal tide energy is lost to the interior ocean internal wave field.
-
real
cdrag¶ The bottom drag coefficient [nondim].
-
logical
apply_background_drag¶ If true, apply a drag due to background processes as a sink.
-
logical
apply_bottom_drag¶ If true, apply a quadratic bottom drag as a sink.
-
logical
apply_wave_drag¶ If true, apply scattering due to small-scale roughness as a sink.
-
logical
apply_froude_drag¶ If true, apply wave breaking as a sink.
-
real, dimension(:,:,:,:,:), pointer mom_internal_tides::int_tide_cs::en => NULL() The internal wave energy density as a function of (i,j,angle,frequency,mode)
-
real, dimension(:,:,:), pointer mom_internal_tides::int_tide_cs::en_restart => NULL() The internal wave energy density as a function of (i,j,angle); temporary for restart.
-
real, dimension(:), allocatable mom_internal_tides::int_tide_cs::frequency The frequency of each band [s-1].
-
type(diag_ctrl), pointer mom_internal_tides::int_tide_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
type(wave_structure_cs), pointer mom_internal_tides::int_tide_cs::wave_structure_csp => NULL() A pointer to the wave_structure module control structure.
-
integer
- type
This control structure holds parameters that regulate internal tide energy inputs.
Unnamed Group
-
integer
id_tke_itidal= -1¶ Diagnostic IDs.
-
integer
id_nb= -1¶ Diagnostic IDs.
-
integer
id_n2_bot= -1¶ Diagnostic IDs.
Public Members
-
logical
debug¶ If true, write verbose checksums for debugging.
-
type(diag_ctrl), pointer mom_int_tide_input::int_tide_input_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
real
tke_itide_max¶ Maximum Internal tide conversion available to mix above the BBL [W m-2].
-
real
kappa_fill¶ Vertical diffusivity used to interpolate sensible values of T & S into thin layers [Z2 T-1 ~> m2 s-1].
-
real, dimension(:,:), allocatable mom_int_tide_input::int_tide_input_cs::tke_itidal_coef The time-invariant field that enters the TKE_itidal input calculation [J m-2].
-
character(len=200) mom_int_tide_input::int_tide_input_cs::inputdir The directory for input files.
-
logical
int_tide_source_test¶ If true, apply an arbitrary generation site for internal tide testing (BDM)
-
type(time_type) mom_int_tide_input::int_tide_input_cs::time_max_source A time for use in testing internal tides.
-
real
int_tide_source_x¶ X Location of generation site for internal tide for testing (BDM)
-
real
int_tide_source_y¶ Y Location of generation site for internal tide for testing (BDM)
-
integer
- type
This type is used to exchange fields related to the internal tides.
Unnamed Group
-
real, dimension(:,:), allocatable mom_int_tide_input::int_tide_input_type::tke_itidal_input The internal tide TKE input at the bottom of the ocean [W m-2].
-
real, dimension(:,:), allocatable mom_int_tide_input::int_tide_input_type::h2 The squared topographic roughness height [Z2 ~> m2].
-
real, dimension(:,:), allocatable mom_int_tide_input::int_tide_input_type::tideamp The amplitude of the tidal velocities [m s-1].
-
real, dimension(:,:), allocatable mom_int_tide_input::int_tide_input_type::nb The bottom stratification [s-1].
-
- type
Control structure for regrid_interp module.
Public Members
-
integer
interpolation_scheme= -1¶ The following parameter is only relevant when used with the target interface densities regridding scheme. It indicates which interpolation to use to determine the grid.
-
logical
boundary_extrapolation¶ Indicate whether high-order boundary extrapolation should be used within boundary cells.
-
integer
-
interface
is_nan¶ Returns .true. if any element of x is a NaN, and .false. otherwise.
Private Functions
-
logical function mom_checksums::is_nan::is_nan_0d(x x) This function returns .true. if x is a NaN, and .false. otherwise.
- Parameters
[in] x: The value to be checked for NaNs.
-
logical function mom_checksums::is_nan::is_nan_1d(x x, skip_mpp skip_mpp) Returns .true. if any element of x is a NaN, and .false. otherwise.
- Parameters
[in] x: The array to be checked for NaNs.[in] skip_mpp: If true, only check this array only on the local PE (default false).
-
logical function mom_checksums::is_nan::is_nan_2d(x x) Returns .true. if any element of x is a NaN, and .false. otherwise.
- Parameters
[in] x: The array to be checked for NaNs.
-
logical function mom_checksums::is_nan::is_nan_3d(x x) Returns .true. if any element of x is a NaN, and .false. otherwise.
- Parameters
[in] x: The array to be checked for NaNs.
-
- type
ISOMIP tracer package control structure.
Public Members
-
logical
coupled_tracers= .false.¶ These tracers are not offered to the coupler.
-
character(len = 200) isomip_tracer::isomip_tracer_cs::tracer_ic_file The full path to the IC file, or ” ” to initialize internally.
-
type(time_type), pointer isomip_tracer::isomip_tracer_cs::time A pointer to the ocean model’s clock.
-
type(tracer_registry_type), pointer isomip_tracer::isomip_tracer_cs::tr_reg => NULL() A pointer to the MOM tracer registry.
-
real, dimension(:,:,:,:), pointer isomip_tracer::isomip_tracer_cs::tr => NULL() The array of tracers used in this package, in g m-3?
-
real, dimension(ntr) isomip_tracer::isomip_tracer_cs::land_val = -1.0 The value of tr used where land is masked out.
-
logical
use_sponge¶ If true, sponges may be applied somewhere in the domain.
-
integer, dimension(ntr) isomip_tracer::isomip_tracer_cs::ind_tr Indices returned by aof_set_coupler_flux if it is used and the surface tracer concentrations are to be provided to the coupler.
-
type(diag_ctrl), pointer isomip_tracer::isomip_tracer_cs::diag A structure that is used to regulate the timing of diagnostic output.
-
type(vardesc), dimension(ntr) isomip_tracer::isomip_tracer_cs::tr_desc Descriptions and metadata for the tracers in this package.
-
logical
- type
This control structure holds the parameters that regulate shear mixing.
Unnamed Group
-
integer
id_kd_shear= -1¶ Diagnostic IDs.
-
integer
id_tke= -1¶ Diagnostic IDs.
-
integer
id_ild2= -1¶ Diagnostic IDs.
-
integer
id_dz_int= -1¶ Diagnostic IDs.
Public Members
-
real
rino_crit¶ The critical shear Richardson number for shear-entrainment. The theoretical value is 0.25. The values found by Jackson et al. are 0.25-0.35.
-
real
shearmix_rate¶ A nondimensional rate scale for shear-driven entrainment. The value given by Jackson et al. is 0.085-0.089.
-
real
fri_curvature¶ A constant giving the curvature of the function of the Richardson number that relates shear to sources in the kappa equation [nondim]. The values found by Jackson et al. are -0.97 - -0.89.
-
real
c_n¶ The coefficient for the decay of TKE due to stratification (i.e. proportional to N*tke) [nondim]. The values found by Jackson et al. are 0.24-0.28.
-
real
c_s¶ The coefficient for the decay of TKE due to shear (i.e. proportional to |S|*tke) [nondim]. The values found by Jackson et al. are 0.14-0.12.
-
real
lambda¶ The coefficient for the buoyancy length scale in the kappa equation. Nondimensional. The values found by Jackson et al. are 0.82-0.81.
-
real
lambda2_n_s¶ The square of the ratio of the coefficients of the buoyancy and shear scales in the diffusivity equation, 0 to eliminate the shear scale. Nondim.
-
real
tke_bg¶ The background level of TKE [Z2 T-2 ~> m2 s-2].
-
real
kappa_0¶ The background diapycnal diffusivity [Z2 T-1 ~> m2 s-1].
-
real
kappa_tol_err¶ The fractional error in kappa that is tolerated.
-
real
prandtl_turb¶ Prandtl number used to convert Kd_shear into viscosity.
-
integer
nkml¶ The number of layers in the mixed layer, as treated in this routine. If the pieces of the mixed layer are not to be treated collectively, nkml is set to 1.
-
integer
max_rino_it¶ The maximum number of iterations that may be used to estimate the instantaneous shear-driven mixing.
-
integer
max_ks_it¶ The maximum number of iterations that may be used to estimate the time-averaged diffusivity.
-
logical
ks_at_vertex¶ If true, do the calculations of the shear-driven mixing at the cell vertices (i.e., the vorticity points).
-
logical
eliminate_massless¶ If true, massless layers are merged with neighboring massive layers in this calculation.
-
real
vel_underflow¶ Velocity components smaller than vel_underflow are set to 0 [Z T-1 ~> m s-1].
-
logical
debug= .false.¶ If true, write verbose debugging messages.
-
type(diag_ctrl), pointer mom_kappa_shear::kappa_shear_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
integer
- type
Control structure for Kelvin wave open boundaries.
Public Members
-
integer
mode= 0¶ Vertical mode.
-
real
coast_angle= 0¶ Angle of coastline.
-
real
coast_offset1= 0¶ Longshore distance to coastal angle.
-
real
coast_offset2= 0¶ Longshore distance to coastal angle.
-
real
h0= 0¶ Bottom depth.
-
real
f_0¶ Coriolis parameter.
-
real
rho_range¶ Density range.
-
real
rho_0¶ Mean density.
-
logical
answers_2018¶ If true, use the order of arithmetic and expressions that recover the answers from the end of 2018. Otherwise, use expressions that give rotational symmetry and eliminate apparent bugs.
-
integer
- type
Control structure for containing KPP parameters/data.
Unnamed Group
-
integer
id_obldepth= -1¶ Diagnostic handles.
-
integer
id_bulkri= -1¶ Diagnostic handles.
-
integer
id_n= -1¶ Diagnostic handles.
-
integer
id_n2= -1¶ Diagnostic handles.
-
integer
id_ws= -1¶ Diagnostic handles.
-
integer
id_vt2= -1¶ Diagnostic handles.
-
integer
id_bulkuz2= -1¶ Diagnostic handles.
-
integer
id_bulkdrho= -1¶ Diagnostic handles.
-
integer
id_ustar= -1¶ Diagnostic handles.
-
integer
id_buoyflux= -1¶ Diagnostic handles.
-
integer
id_qminussw= -1¶ Diagnostic handles.
-
integer
id_nets= -1¶ Diagnostic handles.
-
integer
id_sigma= -1¶ Diagnostic handles.
-
integer
id_kv_kpp= -1¶ Diagnostic handles.
-
integer
id_kt_kpp= -1¶ Diagnostic handles.
-
integer
id_ks_kpp= -1¶ Diagnostic handles.
-
integer
id_tsurf= -1¶ Diagnostic handles.
-
integer
id_ssurf= -1¶ Diagnostic handles.
-
integer
id_usurf= -1¶ Diagnostic handles.
-
integer
id_vsurf= -1¶ Diagnostic handles.
-
integer
id_kd_in= -1¶ Diagnostic handles.
-
integer
id_nltt= -1¶ Diagnostic handles.
-
integer
id_nlts= -1¶ Diagnostic handles.
-
integer
id_nlt_dsdt= -1¶ Diagnostic handles.
-
integer
id_nlt_dtdt= -1¶ Diagnostic handles.
-
integer
id_nlt_temp_budget= -1¶ Diagnostic handles.
-
integer
id_nlt_saln_budget= -1¶ Diagnostic handles.
-
integer
id_enhk= -1¶ Diagnostic handles.
-
integer
id_enhvt2= -1¶ Diagnostic handles.
-
integer
id_enhw= -1¶ Diagnostic handles.
-
integer
id_la_sl= -1¶ Diagnostic handles.
-
integer
id_obldepth_original= -1¶ Diagnostic handles.
-
real, dimension(:,:), allocatable mom_cvmix_kpp::kpp_cs::obldepth Depth (positive) of OBL [m].
-
real, dimension(:,:), allocatable mom_cvmix_kpp::kpp_cs::obldepth_original Depth (positive) of OBL [m] without smoothing.
-
real, dimension(:,:), allocatable mom_cvmix_kpp::kpp_cs::kobl Level (+fraction) of OBL extent.
-
real, dimension(:,:), allocatable mom_cvmix_kpp::kpp_cs::obldepthprev previous Depth (positive) of OBL [m]
-
real, dimension(:,:), allocatable mom_cvmix_kpp::kpp_cs::la_sl Langmuir number used in KPP.
-
real, dimension(:,:,:), allocatable mom_cvmix_kpp::kpp_cs::drho Bulk difference in density [kg m-3].
-
real, dimension(:,:,:), allocatable mom_cvmix_kpp::kpp_cs::uz2 Square of bulk difference in resolved velocity [m2 s-2].
-
real, dimension(:,:,:), allocatable mom_cvmix_kpp::kpp_cs::bulkri Bulk Richardson number for each layer (dimensionless)
-
real, dimension(:,:,:), allocatable mom_cvmix_kpp::kpp_cs::sigma Sigma coordinate (dimensionless)
-
real, dimension(:,:,:), allocatable mom_cvmix_kpp::kpp_cs::ws Turbulent velocity scale for scalars [m s-1].
-
real, dimension(:,:,:), allocatable mom_cvmix_kpp::kpp_cs::n Brunt-Vaisala frequency [s-1].
-
real, dimension(:,:,:), allocatable mom_cvmix_kpp::kpp_cs::n2 Squared Brunt-Vaisala frequency [s-2].
-
real, dimension(:,:,:), allocatable mom_cvmix_kpp::kpp_cs::vt2 Unresolved squared turbulence velocity for bulk Ri [m2 s-2].
-
real, dimension(:,:,:), allocatable mom_cvmix_kpp::kpp_cs::kt_kpp Temp diffusivity from KPP [m2 s-1].
-
real, dimension(:,:,:), allocatable mom_cvmix_kpp::kpp_cs::ks_kpp Scalar diffusivity from KPP [m2 s-1].
-
real, dimension(:,:,:), allocatable mom_cvmix_kpp::kpp_cs::kv_kpp Viscosity due to KPP [m2 s-1].
-
real, dimension(:,:), allocatable mom_cvmix_kpp::kpp_cs::tsurf Temperature of surface layer [degC].
-
real, dimension(:,:), allocatable mom_cvmix_kpp::kpp_cs::ssurf Salinity of surface layer [ppt].
-
real, dimension(:,:), allocatable mom_cvmix_kpp::kpp_cs::usurf i-velocity of surface layer [m s-1]
-
real, dimension(:,:), allocatable mom_cvmix_kpp::kpp_cs::vsurf j-velocity of surface layer [m s-1]
-
real, dimension(:,:,:), allocatable mom_cvmix_kpp::kpp_cs::enhk Enhancement for mixing coefficient.
-
real, dimension(:,:,:), allocatable mom_cvmix_kpp::kpp_cs::enhvt2 Enhancement for Vt2.
Public Members
-
real
ri_crit¶ Critical bulk Richardson number (defines OBL depth)
-
real
vonkarman¶ von Karman constant (dimensionless)
-
real
cs¶ Parameter for computing velocity scale function (dimensionless)
-
real
cs2¶ Parameter for multiplying by non-local term.
-
logical
enhance_diffusion¶ If True, add enhanced diffusivity at base of boundary layer.
-
character(len=10) mom_cvmix_kpp::kpp_cs::interptype Type of interpolation to compute bulk Richardson number.
-
character(len=10) mom_cvmix_kpp::kpp_cs::interptype2 Type of interpolation to compute diff and visc at OBL_depth.
-
logical
computeekman¶ If True, compute Ekman depth limit for OBLdepth.
-
logical
computemoninobukhov¶ If True, compute Monin-Obukhov limit for OBLdepth.
-
logical
passivemode¶ If True, makes KPP passive meaning it does NOT alter the diffusivity.
-
real
deepobloffset¶ If non-zero, is a distance from the bottom that the OBL can not penetrate through [m].
-
real
minobldepth¶ If non-zero, is a minimum depth for the OBL [m].
-
real
surf_layer_ext¶ Fraction of OBL depth considered in the surface layer [nondim].
-
real
minvtsqr¶ Min for the squared unresolved velocity used in Rib CVMix calculation [m2 s-2].
-
logical
fixedobldepth¶ If True, will fix the OBL depth at fixedOBLdepth_value.
-
real
fixedobldepth_value¶ value for the fixed OBL depth when fixedOBLdepth==True.
-
logical
debug¶ If True, calculate checksums and write debugging information.
-
character(len=30) mom_cvmix_kpp::kpp_cs::matchtechnique Method used in CVMix for setting diffusivity and NLT profile functions.
-
integer
nlt_shape¶ MOM6 over-ride of CVMix NLT shape function.
-
logical
applynonlocaltrans¶ If True, apply non-local transport to heat and scalars.
-
integer
n_smooth¶ Number of times smoothing operator is applied on OBLdepth.
-
logical
deepen_only¶ If true, apply OBLdepth smoothing at a cell only if the OBLdepth gets deeper.
-
logical
kppzerodiffusivity¶ If True, will set diffusivity and viscosity from KPP to zero for testing purposes.
-
logical
kppisadditive¶ If True, will add KPP diffusivity to initial diffusivity. If False, will replace initial diffusivity wherever KPP diffusivity is non-zero.
-
real
min_thickness¶ A minimum thickness used to avoid division by small numbers in the vicinity of vanished layers.
-
logical
correctsurflayeravg¶ If true, applies a correction to the averaging of surface layer properties.
-
real
surflayerdepth¶ A guess at the depth of the surface layer (which should 0.1 of OBLdepth) [m].
-
integer
sw_method¶ Sets method for using shortwave radiation in surface buoyancy flux.
-
logical
lt_k_enhancement¶ Flags if enhancing mixing coefficients due to LT.
-
integer
lt_k_shape¶ Integer for constant or shape function enhancement.
-
integer
lt_k_method¶ Integer for mixing coefficients LT method.
-
real
kpp_k_enh_fac¶ Factor to multiply by K if Method is CONSTANT.
-
logical
lt_vt2_enhancement¶ Flags if enhancing Vt2 due to LT.
-
integer
lt_vt2_method¶ Integer for Vt2 LT method.
-
real
kpp_vt2_enh_fac¶ Factor to multiply by VT2 if Method is CONSTANT.
-
logical
stokes_mixing¶ Flag if model is mixing down Stokes gradient This is relavent for which current to use in RiB.
-
type(cvmix_kpp_params_type), pointer mom_cvmix_kpp::kpp_cs::kpp_params => NULL() CVMix parameters.
-
type(diag_ctrl), pointer mom_cvmix_kpp::kpp_cs::diag => NULL() Pointer to diagnostics control structure.
-
integer
- type
A linked list of the parameter documentation messages that have been issued so far.
Private Members
-
type(link_msg), pointer mom_document::link_msg::next => NULL() Facilitates linked list.
-
character(len=80) mom_document::link_msg::name Parameter name.
-
character(len=620) mom_document::link_msg::msg Parameter value and default.
-
- type
A link in the list of variables that have already had override warnings issued.
Unnamed Group
-
type(link_parameter), pointer mom_file_parser::link_parameter::next => NULL() Facilitates linked list.
-
character(len=80) mom_file_parser::link_parameter::name Parameter name.
-
logical
hasissuedoverridewarning= .false.¶ Has a default value.
-
- type
A desciption of the functional dependence of transport at a u-point.
Unnamed Group
-
real
fa_u_ee¶ The effective open face area for zonal barotropic transport drawing from locations far to the east [H L ~> m2 or kg m-1].
-
real
fa_u_e0¶ The effective open face area for zonal barotropic transport drawing from nearby to the east [H L ~> m2 or kg m-1].
-
real
fa_u_w0¶ The effective open face area for zonal barotropic transport drawing from nearby to the west [H L ~> m2 or kg m-1].
-
real
fa_u_ww¶ The effective open face area for zonal barotropic transport drawing from locations far to the west [H L ~> m2 or kg m-1].
-
real
ubt_ww¶ uBT_WW is the barotropic velocity [L T-1 ~> m s-1], beyond which the marginal open face area is FA_u_WW. uBT_WW must be non-negative.
-
real
ubt_ee¶ uBT_EE is a barotropic velocity [L T-1 ~> m s-1], beyond which the marginal open face area is FA_u_EE. uBT_EE must be non-positive.
-
real
uh_crvw¶ The curvature of face area with velocity for flow from the west [H T2 L-1 ~> s2 or kg s2 m-3].
-
real
uh_crve¶ The curvature of face area with velocity for flow from the east [H T2 L-1 ~> s2 or kg s2 m-3].
-
real
uh_ww¶ The zonal transport when ubt=ubt_WW [H L2 T-1 ~> m3 s-1 or kg s-1].
-
real
uh_ee¶ The zonal transport when ubt=ubt_EE [H L2 T-1 ~> m3 s-1 or kg s-1].
-
real
- type
A desciption of the functional dependence of transport at a v-point.
Unnamed Group
-
real
fa_v_nn¶ The effective open face area for meridional barotropic transport drawing from locations far to the north [H m ~> m2 or kg m-1].
-
real
fa_v_n0¶ The effective open face area for meridional barotropic transport drawing from nearby to the north [H m ~> m2 or kg m-1].
-
real
fa_v_s0¶ The effective open face area for meridional barotropic transport drawing from nearby to the south [H m ~> m2 or kg m-1].
-
real
fa_v_ss¶ The effective open face area for meridional barotropic transport drawing from locations far to the south [H m ~> m2 or kg m-1].
-
real
vbt_ss¶ vBT_SS is the barotropic velocity [L T-1 ~> m s-1], beyond which the marginal open face area is FA_v_SS. vBT_SS must be non-negative.
-
real
vbt_nn¶ vBT_NN is the barotropic velocity [L T-1 ~> m s-1], beyond which the marginal open face area is FA_v_NN. vBT_NN must be non-positive.
-
real
vh_crvs¶ The curvature of face area with velocity for flow from the south [H T2 L-1 ~> s2 or kg s2 m-3].
-
real
vh_crvn¶ The curvature of face area with velocity for flow from the north [H T2 L-1 ~> s2 or kg s2 m-3].
-
real
vh_ss¶ The meridional transport when vbt=vbt_SS [H L2 T-1 ~> m3 s-1 or kg s-1].
-
real
vh_nn¶ The meridional transport when vbt=vbt_NN [H L2 T-1 ~> m3 s-1 or kg s-1].
-
real
-
interface
log_param¶ An overloaded interface to log the values of various types of parameters.
Unnamed Group
-
subroutine mom_file_parser::log_param::log_param_int(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, layoutParam layoutParam, debuggingParam debuggingParam) Log the name and value of an integer model parameter in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the module using this parameter[in] varname: The name of the parameter to log[in] value: The value of the parameter to log[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] layoutparam: If present and true, this parameter is logged in the layout parameter file[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file
-
subroutine mom_file_parser::log_param::log_param_real(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, debuggingParam debuggingParam) Log the name and value of a real model parameter in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The name of the parameter to log[in] value: The value of the parameter to log[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file
-
subroutine mom_file_parser::log_param::log_param_logical(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, layoutParam layoutParam, debuggingParam debuggingParam) Log the name and value of a logical model parameter in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The name of the parameter to log[in] value: The value of the parameter to log[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] layoutparam: If present and true, this parameter is logged in the layout parameter file[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file
-
subroutine mom_file_parser::log_param::log_param_char(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, layoutParam layoutParam, debuggingParam debuggingParam) Log the name and value of a character string model parameter in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The name of the parameter to log[in] value: The value of the parameter to log[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] layoutparam: If present and true, this parameter is logged in the layout parameter file[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file
-
subroutine mom_file_parser::log_param::log_param_time(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, timeunit timeunit, layoutParam layoutParam, debuggingParam debuggingParam, log_date log_date) This subroutine writes the value of a time-type parameter to a log file, along with its name and the module it came from.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The name of the parameter to log[in] value: The value of the parameter to log[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] timeunit: The number of seconds in a time unit for real-number output.[in] log_date: If true, log the time_type in date format. If missing the default is false.[in] layoutparam: If present and true, this parameter is logged in the layout parameter file[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file
-
subroutine mom_file_parser::log_param::log_param_int_array(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, layoutParam layoutParam, debuggingParam debuggingParam) Log the name and values of an array of integer model parameter in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the module using this parameter[in] varname: The name of the parameter to log[in] value: The value of the parameter to log[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] layoutparam: If present and true, this parameter is logged in the layout parameter file[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file
-
subroutine mom_file_parser::log_param::log_param_real_array(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, debuggingParam debuggingParam) Log the name and values of an array of real model parameter in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The name of the parameter to log[in] value: The value of the parameter to log[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file
-
-
interface
log_version¶ An overloaded interface to log version information about modules.
Unnamed Group
-
subroutine
log_version_cs(CS CS, modulename modulename, version version, desc desc)¶ Log the version of a module to a log file and/or stdout, and/or to the parameter documentation file.
- Parameters
[in] cs: File parser type[in] modulename: Name of calling module[in] version: Version string of module[in] desc: Module description
-
subroutine
log_version_plain(modulename modulename, version version)¶ Log the version of a module to a log file and/or stdout.
- Parameters
[in] modulename: Name of calling module[in] version: Version string of module
-
subroutine
- type
A structure with the active energy loop bounds.
- type
A container for loop bounds.
- type
Control structure for MOM_marine_ice.
Public Members
-
real
kv_iceberg¶ The viscosity of the icebergs [m2 s-1] (for ice rigidity)
-
real
berg_area_threshold¶ Fraction of grid cell which iceberg must occupy so that fluxes below are set to zero. (0.5 is a good value to use.) Not applied for negative values.
-
real
latent_heat_fusion¶ Latent heat of fusion [J kg-1].
-
real
density_iceberg¶ A typical density of icebergs [kg m-3] (for ice rigidity)
-
type(time_type), pointer mom_marine_ice::marine_ice_cs::time A pointer to the ocean model’s clock.
-
type(diag_ctrl), pointer mom_marine_ice::marine_ice_cs::diag A structure that is used to regulate the timing of diagnostic output.
-
real
- type
Structure that contains pointers to the mechanical forcing at the surface used to drive the liquid ocean simulated by MOM. Data in this type is allocated in the module MOM_surface_forcing.F90, of which there are three versions: solo, coupled, and ice-shelf.
Public Members
-
real, dimension(:,:), pointer mom_forcing_type::mech_forcing::taux => NULL() zonal wind stress [Pa]
-
real, dimension(:,:), pointer mom_forcing_type::mech_forcing::tauy => NULL() meridional wind stress [Pa]
-
real, dimension(:,:), pointer mom_forcing_type::mech_forcing::ustar => NULL() surface friction velocity scale [Z T-1 ~> m s-1].
-
real, dimension(:,:), pointer mom_forcing_type::mech_forcing::net_mass_src => NULL() The net mass source to the ocean [kg m-2 s-1].
-
real, dimension(:,:), pointer mom_forcing_type::mech_forcing::p_surf_full => NULL() Pressure at the top ocean interface [Pa]. if there is sea-ice, then p_surf_flux is at ice-ocean interface.
-
real, dimension(:,:), pointer mom_forcing_type::mech_forcing::p_surf => NULL() Pressure at the top ocean interface [Pa] as used to drive the ocean model. If p_surf is limited, p_surf may be smaller than p_surf_full, otherwise they are the same.
-
real, dimension(:,:), pointer mom_forcing_type::mech_forcing::p_surf_ssh => NULL() Pressure at the top ocean interface that is used in corrections to the sea surface height field that is passed back to the calling routines. p_surf_SSH may point to p_surf or to p_surf_full.
-
real, dimension(:,:), pointer mom_forcing_type::mech_forcing::area_berg => NULL() fractional area of ocean surface covered by icebergs [m2 m-2]
-
real, dimension(:,:), pointer mom_forcing_type::mech_forcing::mass_berg => NULL() mass of icebergs per unit ocean area [kg m-2]
-
real, dimension(:,:), pointer mom_forcing_type::mech_forcing::frac_shelf_u => NULL() Fractional ice shelf coverage of u-cells, nondimensional from 0 to 1 [nondim]. This is only associated if ice shelves are enabled, and is exactly 0 away from shelves or on land.
-
real, dimension(:,:), pointer mom_forcing_type::mech_forcing::frac_shelf_v => NULL() Fractional ice shelf coverage of v-cells, nondimensional from 0 to 1 [nondim]. This is only associated if ice shelves are enabled, and is exactly 0 away from shelves or on land.
-
real, dimension(:,:), pointer mom_forcing_type::mech_forcing::rigidity_ice_u => NULL() Depth-integrated lateral viscosity of ice shelves or sea ice at u-points [m3 s-1].
-
real, dimension(:,:), pointer mom_forcing_type::mech_forcing::rigidity_ice_v => NULL() Depth-integrated lateral viscosity of ice shelves or sea ice at v-points [m3 s-1].
-
real
dt_force_accum= -1.0¶ The amount of time over which the mechanical forcing fluxes have been averaged [s].
-
logical
net_mass_src_set= .false.¶ If true, an estimate of net_mass_src has been provided.
-
logical
accumulate_p_surf= .false.¶ If true, the surface pressure due to the atmosphere and various types of ice needs to be accumulated, and the surface pressure explicitly reset to zero at the driver level when appropriate.
-
logical
accumulate_rigidity= .false.¶ If true, the rigidity due to various types of ice needs to be accumulated, and the rigidity explicitly reset to zero at the driver level when appropriate.
-
logical
initialized= .false.¶ This indicates whether the appropriate arrays have been initialized.
-
- type
Control structure that contains MEKE parameters and diagnostics handles.
Unnamed Group
-
integer
id_meke= -1¶ Diagnostic handles.
-
integer
id_ue= -1¶ Diagnostic handles.
-
integer
id_kh= -1¶ Diagnostic handles.
-
integer
id_src= -1¶ Diagnostic handles.
-
integer
id_ub= -1¶ Diagnostic handles.
-
integer
id_ut= -1¶ Diagnostic handles.
-
integer
id_gm_src= -1¶ Diagnostic handles.
-
integer
id_mom_src= -1¶ Diagnostic handles.
-
integer
id_gme_snk= -1¶ Diagnostic handles.
-
integer
id_decay= -1¶ Diagnostic handles.
-
integer
id_khmeke_u= -1¶ Diagnostic handles.
-
integer
id_khmeke_v= -1¶ Diagnostic handles.
-
integer
id_ku= -1¶ Diagnostic handles.
-
integer
id_au= -1¶ Diagnostic handles.
-
integer
id_le= -1¶ Diagnostic handles.
-
integer
id_gamma_b= -1¶ Diagnostic handles.
-
integer
id_gamma_t= -1¶ Diagnostic handles.
-
integer
id_lrhines= -1¶ Diagnostic handles.
-
integer
id_leady= -1¶ Diagnostic handles.
-
integer
id_clock_pass¶ Clock for group pass calls.
-
type(group_pass_type) mom_meke::meke_cs::pass_meke Group halo pass handle for MEKEMEKE and maybe MEKEKh_diff.
-
type(group_pass_type) mom_meke::meke_cs::pass_kh Group halo pass handle for MEKEKh, MEKEKu, and/or MEKEAu.
Public Members
-
real
meke_frcoeff¶ Efficiency of conversion of ME into MEKE [nondim].
-
real
meke_gmcoeff¶ Efficiency of conversion of PE into MEKE [nondim].
-
real
meke_gmecoeff¶ Efficiency of conversion of MEKE into ME by GME [nondim].
-
real
meke_damping¶ Local depth-independent MEKE dissipation rate [s-1].
-
real
meke_cd_scale¶ The ratio of the bottom eddy velocity to the column mean eddy velocity, i.e. sqrt(2*MEKE). This should be less than 1 to account for the surface intensification of MEKE.
-
real
meke_cb¶ Coefficient in the \(\gamma_{bot}\) expression [nondim].
-
real
meke_min_gamma¶ Minimum value of gamma_b^2 allowed [nondim].
-
real
meke_ct¶ Coefficient in the \(\gamma_{bt}\) expression [nondim].
-
logical
visc_drag¶ If true use the vertvisc_type to calculate bottom drag.
-
logical
jansen15_drag¶ If true use the bottom drag formulation from Jansen et al. (2015)
-
logical
meke_geometric¶ If true, uses the GM coefficient formulation from the GEOMETRIC framework (Marshall et al., 2012)
-
logical
gm_src_alt¶ If true, use the GM energy conversion form S^2*N^2*kappa rather than the streamfunction for the MEKE GM source term.
-
logical
rd_as_max_scale¶ If true the length scale can not exceed the first baroclinic deformation radius.
-
logical
use_old_lscale¶ Use the old formula for mixing length scale.
-
logical
use_min_lscale¶ Use simple minimum for mixing length scale.
-
real
cdrag¶ The bottom drag coefficient for MEKE [nondim].
-
real
meke_bgsrc¶ Background energy source for MEKE [W kg-1] (= m2 s-3).
-
real
meke_dtscale¶ Scale factor to accelerate time-stepping [nondim].
-
real
meke_khcoeff¶ Scaling factor to convert MEKE into Kh [nondim].
-
real
meke_uscale¶ MEKE velocity scale for bottom drag [m s-1].
-
real
meke_kh¶ Background lateral diffusion of MEKE [m2 s-1].
-
real
meke_k4¶ Background bi-harmonic diffusivity (of MEKE) [m4 s-1].
-
real
khmeke_fac¶ A factor relating MEKEKh to the diffusivity used for MEKE itself [nondim].
-
real
viscosity_coeff_ku¶ The scaling coefficient in the expression for viscosity used to parameterize lateral harmonic momentum mixing by unresolved eddies represented by MEKE.
-
real
viscosity_coeff_au¶ The scaling coefficient in the expression for viscosity used to parameterize lateral biharmonic momentum mixing by unresolved eddies represented by MEKE.
-
real
lfixed¶ Fixed mixing length scale [m].
-
real
adeform¶ Weighting towards deformation scale of mixing length [nondim].
-
real
arhines¶ Weighting towards Rhines scale of mixing length [nondim].
-
real
africt¶ Weighting towards frictional arrest scale of mixing length [nondim].
-
real
aeady¶ Weighting towards Eady scale of mixing length [nondim].
-
real
agrid¶ Weighting towards grid scale of mixing length [nondim].
-
real
meke_advection_factor¶ A scaling in front of the advection of MEKE [nondim].
-
real
meke_topographic_beta¶ Weight for how much topographic beta is considered when computing beta in Rhines scale [nondim].
-
logical
kh_flux_enabled¶ If true, lateral diffusive MEKE flux is enabled.
-
logical
initialize¶ If True, invokes a steady state solver to calculate MEKE.
-
logical
debug¶ If true, write out checksums of data for debugging.
-
type(diag_ctrl), pointer mom_meke::meke_cs::diag => NULL() A type that regulates diagnostics output.
-
integer
- type
This type is used to exchange information related to the MEKE calculations.
Public Members
-
real, dimension(:,:), pointer mom_meke_types::meke_type::meke => NULL() Vertically averaged eddy kinetic energy [m2 s-2].
-
real, dimension(:,:), pointer mom_meke_types::meke_type::gm_src => NULL() MEKE source due to thickness mixing (GM) [W m-2].
-
real, dimension(:,:), pointer mom_meke_types::meke_type::mom_src => NULL() MEKE source from lateral friction in the momentum equations [W m-2].
-
real, dimension(:,:), pointer mom_meke_types::meke_type::gme_snk => NULL() MEKE sink from GME backscatter in the momentum equations [W m-2].
-
real, dimension(:,:), pointer mom_meke_types::meke_type::kh => NULL() The MEKE-derived lateral mixing coefficient [m2 s-1].
-
real, dimension(:,:), pointer mom_meke_types::meke_type::kh_diff => NULL() Uses the non-MEKE-derived thickness diffusion coefficient to diffuse.
-
real, dimension(:,:), pointer mom_meke_types::meke_type::rd_dx_h => NULL() The deformation radius compared with the grid spacing [nondim].
-
real, dimension(:,:), pointer mom_meke_types::meke_type::ku => NULL() The MEKE-derived lateral viscosity coefficient [m2 T-1 ~> m2 s-1]. This viscosity can be negative when representing backscatter from unresolved eddies (see Jansen and Held, 2014).
-
real, dimension(:,:), pointer mom_meke_types::meke_type::au => NULL() The MEKE-derived lateral biharmonic viscosity coefficient [m4 T-1 ~> m4 s-1].
-
real
khth_fac= 1.0¶ Multiplier to map Kh(MEKE) to KhTh [nondim].
-
real
khtr_fac= 1.0¶ Multiplier to map Kh(MEKE) to KhTr [nondim].
-
real
backscatter_ro_pow= 0.0¶ Power in Rossby number function for backscatter.
-
real
backscatter_ro_c= 0.0¶ Coefficient in Rossby number function for backscatter.
-
- type
A container for passing around active tracer point memory limits.
- type
This control structure is used to store parameters associated with the MESO forcing.
Public Members
-
logical
use_temperature¶ If true, temperature and salinity are used as state variables.
-
logical
restorebuoy¶ If true, use restoring surface buoyancy forcing.
-
real
rho0¶ The density used in the Boussinesq approximation [kg m-3].
-
real
g_earth¶ The gravitational acceleration [L2 Z-1 T-2 ~> m s-2].
-
real
flux_const¶ The restoring rate at the surface [m s-1].
-
real
gust_const¶ A constant unresolved background gustiness that contributes to ustar [Pa].
-
real, dimension(:,:), pointer meso_surface_forcing::meso_surface_forcing_cs::t_restore => NULL() The temperature to restore the SST toward [degC].
-
real, dimension(:,:), pointer meso_surface_forcing::meso_surface_forcing_cs::s_restore => NULL() The salinity to restore the sea surface salnity toward [ppt].
-
real, dimension(:,:), pointer meso_surface_forcing::meso_surface_forcing_cs::pme => NULL() The prescribed precip minus evap [m s-1].
-
real, dimension(:,:), pointer meso_surface_forcing::meso_surface_forcing_cs::solar => NULL() The shortwave forcing into the ocean [W m-2].
-
real, dimension(:,:), pointer meso_surface_forcing::meso_surface_forcing_cs::heat => NULL() The prescribed longwave, latent and sensible heat flux into the ocean [W m-2].
-
character(len=200) meso_surface_forcing::meso_surface_forcing_cs::inputdir The directory where NetCDF input files are.
-
character(len=200) meso_surface_forcing::meso_surface_forcing_cs::salinityrestore_file The file with the target sea surface salinity.
-
character(len=200) meso_surface_forcing::meso_surface_forcing_cs::sstrestore_file The file with the target sea surface temperature.
-
character(len=200) meso_surface_forcing::meso_surface_forcing_cs::solar_file The file with the shortwave forcing.
-
character(len=200) meso_surface_forcing::meso_surface_forcing_cs::heating_file The file with the longwave, latent, and sensible heating.
-
character(len=200) meso_surface_forcing::meso_surface_forcing_cs::pme_file The file with precipitation minus evaporation.
-
type(diag_ctrl), pointer meso_surface_forcing::meso_surface_forcing_cs::diag A structure that is used to regulate the timing of diagnostic output.
-
logical
- type
Control structure for mom_mixed_layer_restrat.
Unnamed Group
-
integer
id_urestrat_time= -1¶ Diagnostic identifier.
-
integer
id_vrestrat_time= -1¶ Diagnostic identifier.
-
integer
id_uhml= -1¶ Diagnostic identifier.
-
integer
id_vhml= -1¶ Diagnostic identifier.
-
integer
id_mld= -1¶ Diagnostic identifier.
-
integer
id_rml= -1¶ Diagnostic identifier.
-
integer
id_udml= -1¶ Diagnostic identifier.
-
integer
id_vdml= -1¶ Diagnostic identifier.
-
integer
id_uml= -1¶ Diagnostic identifier.
-
integer
id_vml= -1¶ Diagnostic identifier.
Public Members
-
real
ml_restrat_coef¶ A non-dimensional factor by which the instability is enhanced over what would be predicted based on the resolved gradients [nondim]. This increases with grid spacing^2, up to something of order 500.
-
real
ml_restrat_coef2¶ As for ml_restrat_coef but using the slow filtered MLD [nondim].
-
real
front_length¶ If non-zero, is the frontal-length scale [m] used to calculate the upscaling of buoyancy gradients that is otherwise represented by the parameter FOX_KEMPER_ML_RESTRAT_COEF. If MLE_FRONT_LENGTH is non-zero, it is recommended to set FOX_KEMPER_ML_RESTRAT_COEF=1.0.
-
logical
mle_use_pbl_mld¶ If true, use the MLD provided by the PBL parameterization. if false, MLE will calculate a MLD based on a density difference based on the parameter MLE_DENSITY_DIFF.
-
real
mle_mld_decay_time¶ Time-scale to use in a running-mean when MLD is retreating [s].
-
real
mle_mld_decay_time2¶ Time-scale to use in a running-mean when filtered MLD is retreating [s].
-
real
mle_density_diff¶ Density difference used in detecting mixed-layer depth [kgm-3].
-
real
mle_tail_dh¶ Fraction by which to extend the mixed-layer restratification depth used for a smoother stream function at the base of the mixed-layer [nondim].
-
real
mle_mld_stretch¶ A scaling coefficient for stretching/shrinking the MLD used in the MLE scheme [nondim]. This simply multiplies MLD wherever used.
-
logical
mle_use_mld_ave_bug¶ If true, do not account for MLD mismatch to interface positions.
-
logical
debug= .false.¶ If true, calculate checksums of fields for debugging.
-
type(diag_ctrl), pointer mom_mixed_layer_restrat::mixedlayer_restrat_cs::diag A structure that is used to regulate the timing of diagnostic output.
-
real, dimension(:,:), pointer mom_mixed_layer_restrat::mixedlayer_restrat_cs::mld_filtered => NULL() Time-filtered MLD [H ~> m or kg m-2].
-
real, dimension(:,:), pointer mom_mixed_layer_restrat::mixedlayer_restrat_cs::mld_filtered_slow => NULL() Slower time-filtered MLD [H ~> m or kg m-2].
-
integer
- type
Control structure for the MOM module, including the variables that describe the state of the ocean.
Unnamed Group
-
real, dimension( : , : , : ), allocatable mom::mom_control_struct::h layer thickness [H ~> m or kg m-2]
-
real, dimension( : , : , : ), allocatable mom::mom_control_struct::t potential temperature [degC]
-
real, dimension( : , : , : ), allocatable mom::mom_control_struct::s salinity [ppt]
-
real, dimension( : , : , : ), allocatable mom::mom_control_struct::u zonal velocity component [m s-1]
-
real, dimension( : , : , : ), allocatable mom::mom_control_struct::uh uh = u * h * dy at u grid points [H m2 s-1 ~> m3 s-1 or kg s-1]
-
real, dimension( : , : , : ), allocatable mom::mom_control_struct::uhtr accumulated zonal thickness fluxes to advect tracers [H m2 ~> m3 or kg]
-
real, dimension( : , : , : ), allocatable mom::mom_control_struct::v meridional velocity [m s-1]
-
real, dimension( : , : , : ), allocatable mom::mom_control_struct::vh vh = v * h * dx at v grid points [H m2 s-1 ~> m3 s-1 or kg s-1]
-
real, dimension( : , : , : ), allocatable mom::mom_control_struct::vhtr accumulated meridional thickness fluxes to advect tracers [H m2 ~> m3 or kg]
-
real, dimension( : , : ), allocatable mom::mom_control_struct::ssh_rint A running time integral of the sea surface height [s m].
-
real, dimension( : , : ), allocatable mom::mom_control_struct::ave_ssh_ibc time-averaged (over a forcing time step) sea surface height with a correction for the inverse barometer [m]
-
real, dimension( : , : ), allocatable mom::mom_control_struct::eta_av_bc free surface height or column mass time averaged over the last baroclinic dynamics time step [H ~> m or kg m-2]
-
real, dimension(:,:), pointer mom::mom_control_struct::hml => NULL() active mixed layer depth [m]
-
real
time_in_cycle¶ The running time of the current time-stepping cycle in calls that step the dynamics, and also the length of the time integral of ssh_rint [s].
-
real
time_in_thermo_cycle¶ The running time of the current time-stepping cycle in calls that step the thermodynamics [s].
-
type(ocean_grid_type) mom::mom_control_struct::g structure containing metrics and grid info
-
type(verticalgrid_type), pointer mom::mom_control_struct::gv => NULL() structure containing vertical grid info
-
type(unit_scale_type), pointer mom::mom_control_struct::us => NULL() structure containing various unit conversion factors
-
type(thermo_var_ptrs) mom::mom_control_struct::tv structure containing pointers to available thermodynamic fields
-
real
t_dyn_rel_adv¶ The time of the dynamics relative to tracer advection and lateral mixing (in seconds), or equivalently the elapsed time since advectively updating the tracers. t_dyn_rel_adv is invariably positive and may span multiple coupling timesteps.
-
real
t_dyn_rel_thermo¶ The time of the dynamics relative to diabatic processes and remapping (in seconds). t_dyn_rel_thermo can be negative or positive depending on whether the diabatic processes are applied before or after the dynamics and may span multiple coupling timesteps.
-
real
t_dyn_rel_diag¶ The time of the diagnostics relative to diabatic processes and remapping (in seconds). t_dyn_rel_diag is always positive, since the diagnostics must lag.
-
integer
ndyn_per_adv= 0¶ Number of calls to dynamics since the last call to advection.
-
type(diag_ctrl) mom::mom_control_struct::diag structure to regulate diagnostic output timing
-
type(vertvisc_type) mom::mom_control_struct::visc structure containing vertical viscosities, bottom drag viscosities, and related fields
-
type(meke_type), pointer mom::mom_control_struct::meke => NULL() structure containing fields related to the Mesoscale Eddy Kinetic Energy
-
logical
adiabatic¶ If true, there are no diapycnal mass fluxes, and no calls to routines to calculate or apply diapycnal fluxes.
-
logical
diabatic_first¶ If true, apply diabatic and thermodynamic processes before time stepping the dynamics.
-
logical
use_ale_algorithm¶ If true, use the ALE algorithm rather than layered isopycnal/stacked shallow water mode. This logical is set by calling the function useRegridding() from the MOM_regridding module.
-
logical
offline_tracer_mode= .false.¶ If true, step_offline() is called instead of step_MOM(). This is intended for running MOM6 in offline tracer mode.
-
type(time_type), pointer mom::mom_control_struct::time pointer to the ocean clock
-
real
dt¶ (baroclinic) dynamics time step [s]
-
real
dt_therm¶ thermodynamics time step [s]
-
logical
thermo_spans_coupling¶ If true, thermodynamic and tracer time steps can span multiple coupled time steps.
-
integer
nstep_tot= 0¶ The total number of dynamic timesteps tcaaken so far in this run segment.
-
logical
count_calls= .false.¶ If true, count the calls to step_MOM, rather than the number of dynamics steps in nstep_tot.
-
logical
debug¶ If true, write verbose checksums for debugging purposes.
-
integer
ntrunc¶ number u,v truncations since last call to write_energy
-
logical
do_dynamics¶ If false, does not call step_MOM_dyn_*. This is an undocumented run-time flag that is fragile.
-
logical
split¶ If true, use the split time stepping scheme.
-
logical
use_rk2¶ If true, use RK2 instead of RK3 in unsplit mode (i.e., no split between barotropic and baroclinic).
-
logical
thickness_diffuse¶ If true, diffuse interface height w/ a diffusivity KHTH.
-
logical
thickness_diffuse_first¶ If true, diffuse thickness before dynamics.
-
logical
mixedlayer_restrat¶ If true, use submesoscale mixed layer restratifying scheme.
-
logical
usemeke¶ If true, call the MEKE parameterization.
-
logical
usewaves¶ If true, update Stokes drift.
-
real
dtbt_reset_period¶ The time interval between dynamic recalculation of the barotropic time step [s]. If this is negative dtbt is never calculated, and if it is 0, dtbt is calculated every step.
-
type(time_type) mom::mom_control_struct::dtbt_reset_interval A time_time representation of dtbt_reset_period.
-
type(time_type) mom::mom_control_struct::dtbt_reset_time The next time DTBT should be calculated.
-
real, dimension(:,:,:), pointer mom::mom_control_struct::h_pre_dyn => NULL() The thickness before the transports [H ~> m or kg m-2].
-
real, dimension(:,:,:), pointer mom::mom_control_struct::t_pre_dyn => NULL() Temperature before the transports [degC].
-
real, dimension(:,:,:), pointer mom::mom_control_struct::s_pre_dyn => NULL() Salinity before the transports [ppt].
-
type(accel_diag_ptrs) mom::mom_control_struct::adp structure containing pointers to accelerations, for derived diagnostics (e.g., energy budgets)
-
type(cont_diag_ptrs) mom::mom_control_struct::cdp structure containing pointers to continuity equation terms, for derived diagnostics (e.g., energy budgets)
-
real, dimension(:,:,:), pointer mom::mom_control_struct::u_prev => NULL() previous value of u stored for diagnostics [m s-1]
-
real, dimension(:,:,:), pointer mom::mom_control_struct::v_prev => NULL() previous value of v stored for diagnostics [m s-1]
-
logical
interp_p_surf¶ If true, linearly interpolate surface pressure over the coupling time step, using specified value at the end of the coupling step. False by default.
-
logical
p_surf_prev_set¶ If true, p_surf_prev has been properly set from a previous time-step or the ocean restart file. This is only valid when interp_p_surf is true.
-
real, dimension(:,:), pointer mom::mom_control_struct::p_surf_prev => NULL() surface pressure [Pa] at end previous call to step_MOM
-
real, dimension(:,:), pointer mom::mom_control_struct::p_surf_begin => NULL() surface pressure [Pa] at start of step_MOM_dyn_…
-
real, dimension(:,:), pointer mom::mom_control_struct::p_surf_end => NULL() surface pressure [Pa] at end of step_MOM_dyn_…
-
logical
write_ic¶ If true, then the initial conditions will be written to file.
-
character(len=120) mom::mom_control_struct::ic_file A file into which the initial conditions are written in a new run if SAVE_INITIAL_CONDS is true.
-
logical
calc_rho_for_sea_lev¶ If true, calculate rho to convert pressure to sea level.
-
real
hmix¶ Diagnostic mixed layer thickness over which to average surface tracer properties when a bulk mixed layer is not used [Z ~> m], or a negative value if a bulk mixed layer is being used.
-
real
hfrz¶ If HFrz > 0, melt potential will be computed. The actual depth over which melt potential is computed will min(HFrz, OBLD), where OBLD is the boundary layer depth. If HFrz <= 0 (default), melt potential will not be computed.
-
real
hmix_uv¶ Depth scale over which to average surface flow to feedback to the coupler/driver [Z ~> m] when bulk mixed layer is not used, or a negative value if a bulk mixed layer is being used.
-
logical
check_bad_sfc_vals¶ If true, scan surface state for ridiculous values.
-
real
bad_val_ssh_max¶ Maximum SSH before triggering bad value message [m].
-
real
bad_val_sst_max¶ Maximum SST before triggering bad value message [degC].
-
real
bad_val_sst_min¶ Minimum SST before triggering bad value message [degC].
-
real
bad_val_sss_max¶ Maximum SSS before triggering bad value message [ppt].
-
real
bad_val_col_thick¶ Minimum column thickness before triggering bad value message [m].
-
type(mom_diag_ids) mom::mom_control_struct::ids Handles used for diagnostics.
-
type(transport_diag_ids) mom::mom_control_struct::transport_ids Handles used for transport diagnostics.
-
type(surface_diag_ids) mom::mom_control_struct::sfc_ids Handles used for surface diagnostics.
-
type(diag_grid_storage) mom::mom_control_struct::diag_pre_sync The grid (thicknesses) before remapping.
-
type(diag_grid_storage) mom::mom_control_struct::diag_pre_dyn The grid (thicknesses) before dynamics.
-
type(mom_dyn_unsplit_cs), pointer mom::mom_control_struct::dyn_unsplit_csp => NULL() Pointer to the control structure used for the unsplit dynamics.
-
type(mom_dyn_unsplit_rk2_cs), pointer mom::mom_control_struct::dyn_unsplit_rk2_csp => NULL() Pointer to the control structure used for the unsplit RK2 dynamics.
-
type(mom_dyn_split_rk2_cs), pointer mom::mom_control_struct::dyn_split_rk2_csp => NULL() Pointer to the control structure used for the mode-split RK2 dynamics.
-
type(thickness_diffuse_cs), pointer mom::mom_control_struct::thickness_diffuse_csp => NULL() Pointer to the control structure used for the isopycnal height diffusive transport. This is also common referred to as Gent-McWilliams diffusion.
-
type(mixedlayer_restrat_cs), pointer mom::mom_control_struct::mixedlayer_restrat_csp => NULL() Pointer to the control structure used for the mixed layer restratification.
-
type(set_visc_cs), pointer mom::mom_control_struct::set_visc_csp => NULL() Pointer to the control structure used to set viscosities.
-
type(diabatic_cs), pointer mom::mom_control_struct::diabatic_csp => NULL() Pointer to the control structure for the diabatic driver.
-
type(meke_cs), pointer mom::mom_control_struct::meke_csp => NULL() Pointer to the control structure for the MEKE updates.
-
type(varmix_cs), pointer mom::mom_control_struct::varmix => NULL() Pointer to the control structure for the variable mixing module.
-
type(barotropic_cs), pointer mom::mom_control_struct::barotropic_csp => NULL() Pointer to the control structure for the barotropic module.
-
type(tracer_registry_type), pointer mom::mom_control_struct::tracer_reg => NULL() Pointer to the MOM tracer registry.
-
type(tracer_advect_cs), pointer mom::mom_control_struct::tracer_adv_csp => NULL() Pointer to the MOM tracer advection control structure.
-
type(tracer_hor_diff_cs), pointer mom::mom_control_struct::tracer_diff_csp => NULL() Pointer to the MOM along-isopycnal tracer diffusion control structure.
-
type(tracer_flow_control_cs), pointer mom::mom_control_struct::tracer_flow_csp => NULL() Pointer to the control structure that orchestrates the calling of tracer packages.
-
type(update_obc_cs), pointer mom::mom_control_struct::update_obc_csp => NULL() Pointer to the control structure for updating open boundary condition properties.
-
type(ocean_obc_type), pointer mom::mom_control_struct::obc => NULL() Pointer to the MOM open boundary condition type.
-
type(sponge_cs), pointer mom::mom_control_struct::sponge_csp => NULL() Pointer to the layered-mode sponge control structure.
-
type(ale_sponge_cs), pointer mom::mom_control_struct::ale_sponge_csp => NULL() Pointer to the ALE-mode sponge control structure.
-
type(ale_cs), pointer mom::mom_control_struct::ale_csp => NULL() Pointer to the Arbitrary Lagrangian Eulerian (ALE) vertical coordinate control structure.
-
type(sum_output_cs), pointer mom::mom_control_struct::sum_output_csp => NULL() Pointer to the globally summed output control structure.
-
type(diagnostics_cs), pointer mom::mom_control_struct::diagnostics_csp => NULL() Pointer to the MOM diagnostics control structure.
-
type(offline_transport_cs), pointer mom::mom_control_struct::offline_csp => NULL() Pointer to the offline tracer transport control structure.
-
logical
ensemble_ocean¶ if true, this run is part of a larger ensemble for the purpose of data assimilation or statistical analysis.
-
type(oda_cs), pointer mom::mom_control_struct::odacs => NULL() a pointer to the control structure for handling ensemble model state vectors and data assimilation increments and priors
-
- type
A structure with diagnostic IDs of the state variables.
- type
The MOM_domain_type contains information about the domain decompositoin.
Public Members
-
type(domain2d), pointer mom_domains::mom_domain_type::mpp_domain => NULL() The FMS domain with halos on this processor, centered at h points.
-
type(domain2d), pointer mom_domains::mom_domain_type::mpp_domain_d2 => NULL() A coarse FMS domain with halos on this processor, centered at h points.
-
integer
niglobal¶ The total horizontal i-domain size.
-
integer
njglobal¶ The total horizontal j-domain size.
-
integer
nihalo¶ The i-halo size in memory.
-
integer
njhalo¶ The j-halo size in memory.
-
logical
symmetric¶ True if symmetric memory is used with this domain.
-
logical
nonblocking_updates¶ If true, non-blocking halo updates are allowed. The default is .false. (for now).
-
logical
thin_halo_updates¶ If true, optional arguments may be used to specify the width of the halos that are updated with each call.
-
integer, dimension(2) mom_domains::mom_domain_type::layout This domain’s processor layout. This is saved to enable the construction of related new domains with different resolutions or other properties.
-
integer, dimension(2) mom_domains::mom_domain_type::io_layout The IO-layout used with this domain.
-
integer
x_flags¶ Flag that specifies the properties of the domain in the i-direction in a define_domain call.
-
integer
y_flags¶ Flag that specifies the properties of the domain in the j-direction in a define_domain call.
-
logical, dimension(:,:), pointer mom_domains::mom_domain_type::maskmap => NULL() A pointer to an array indicating which logical processors are actually used for the ocean code. The other logical processors would be contain only land points and are not assigned to actual processors. This need not be assigned if all logical processors are used.
-
- type
MOM_dynamics_split_RK2 module control structure.
Unnamed Group
-
integer
id_uh= -1¶ Diagnostic IDs.
-
integer
id_vh= -1¶ Diagnostic IDs.
-
integer
id_umo= -1¶ Diagnostic IDs.
-
integer
id_vmo= -1¶ Diagnostic IDs.
-
integer
id_umo_2d= -1¶ Diagnostic IDs.
-
integer
id_vmo_2d= -1¶ Diagnostic IDs.
-
integer
id_pfu= -1¶ Diagnostic IDs.
-
integer
id_pfv= -1¶ Diagnostic IDs.
-
integer
id_cau= -1¶ Diagnostic IDs.
-
integer
id_cav= -1¶ Diagnostic IDs.
-
integer
id_uav= -1¶ Diagnostic IDs.
-
integer
id_vav= -1¶ Diagnostic IDs.
-
integer
id_u_bt_accel= -1¶ Diagnostic IDs.
-
integer
id_v_bt_accel= -1¶ Diagnostic IDs.
-
type(diag_ctrl), pointer mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::diag A structure that is used to regulate the timing of diagnostic output.
-
type(accel_diag_ptrs), pointer mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::adp A structure pointing to the various accelerations in the momentum equations, which can later be used to calculate derived diagnostics like energy budgets.
-
type(cont_diag_ptrs), pointer mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::cdp A structure with pointers to various terms in the continuity equations, which can later be used to calculate derived diagnostics like energy budgets.
-
type(hor_visc_cs), pointer mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::hor_visc_csp => NULL() A pointer to the horizontal viscosity control structure.
-
type(continuity_cs), pointer mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::continuity_csp => NULL() A pointer to the continuity control structure.
-
type(coriolisadv_cs), pointer mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::coriolisadv_csp => NULL() A pointer to the CoriolisAdv control structure.
-
type(pressureforce_cs), pointer mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::pressureforce_csp => NULL() A pointer to the PressureForce control structure.
-
type(barotropic_cs), pointer mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::barotropic_csp => NULL() A pointer to the barotropic stepping control structure.
-
type(thickness_diffuse_cs), pointer mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::thickness_diffuse_csp => NULL() A pointer to a structure containing interface height diffusivities.
-
type(vertvisc_cs), pointer mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::vertvisc_csp => NULL() A pointer to the vertical viscosity control structure.
-
type(set_visc_cs), pointer mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::set_visc_csp => NULL() A pointer to the set_visc control structure.
-
type(tidal_forcing_cs), pointer mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::tides_csp => NULL() A pointer to the tidal forcing control structure.
-
type(ale_cs), pointer mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::ale_csp => NULL() A pointer to the ALE control structure.
-
type(ocean_obc_type), pointer mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::obc => NULL() A pointer to an open boundary condition type that specifies whether, where, and what open boundary conditions are used. If no open BCs are used, this pointer stays nullified. Flather OBCs use open boundary_CS as well.
-
type(update_obc_cs), pointer mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::update_obc_csp => NULL() A pointer to the update_OBC control structure.
-
type(group_pass_type) mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::pass_eta Structure for group halo pass.
-
type(group_pass_type) mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::pass_visc_rem Structure for group halo pass.
-
type(group_pass_type) mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::pass_uvp Structure for group halo pass.
-
type(group_pass_type) mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::pass_hp_uv Structure for group halo pass.
-
type(group_pass_type) mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::pass_uv Structure for group halo pass.
-
type(group_pass_type) mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::pass_h Structure for group halo pass.
-
type(group_pass_type) mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::pass_av_uvh Structure for group halo pass.
Public Members
-
real, dimension( : , : , : ), allocatable mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::cau CAu = f*v - u.grad(u) [m s-2].
-
real, dimension( : , : , : ), allocatable mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::pfu PFu = -dM/dx [m s-2].
-
real, dimension( : , : , : ), allocatable mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::diffu Zonal acceleration due to convergence of the along-isopycnal stress tensor [m s-1 T-1 ~> m s-2].
-
real, dimension( : , : , : ), allocatable mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::cav CAv = -f*u - u.grad(v) [m s-2].
-
real, dimension( : , : , : ), allocatable mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::pfv PFv = -dM/dy [m s-2].
-
real, dimension( : , : , : ), allocatable mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::diffv Meridional acceleration due to convergence of the along-isopycnal stress tensor [m s-1 T-1 ~> m s-2].
-
real, dimension( : , : , : ), allocatable mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::visc_rem_u Both the fraction of the zonal momentum originally in a layer that remains after a time-step of viscosity, and the fraction of a time-step worth of a barotropic acceleration that a layer experiences after viscosity is applied. Nondimensional between 0 (at the bottom) and 1 (far above).
-
real, dimension( : , : , : ), allocatable mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::u_accel_bt The zonal layer accelerations due to the difference between the barotropic accelerations and the baroclinic accelerations that were fed into the barotopic calculation [m s-2].
-
real, dimension( : , : , : ), allocatable mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::visc_rem_v Both the fraction of the meridional momentum originally in a layer that remains after a time-step of viscosity, and the fraction of a time-step worth of a barotropic acceleration that a layer experiences after viscosity is applied. Nondimensional between 0 (at the bottom) and 1 (far above).
-
real, dimension( : , : , : ), allocatable mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::v_accel_bt The meridional layer accelerations due to the difference between the barotropic accelerations and the baroclinic accelerations that were fed into the barotopic calculation [m s-2].
-
real, dimension( : , : ), allocatable mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::eta Instantaneous free surface height (in Boussinesq mode) or column mass anomaly (in non-Boussinesq mode) [H ~> m or kg m-2].
-
real, dimension( : , : , : ), allocatable mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::u_av layer x-velocity with vertical mean replaced by time-mean barotropic velocity over a baroclinic timestep [m s-1]
-
real, dimension( : , : , : ), allocatable mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::v_av layer y-velocity with vertical mean replaced by time-mean barotropic velocity over a baroclinic timestep [m s-1]
-
real, dimension( : , : , : ), allocatable mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::h_av arithmetic mean of two successive layer thicknesses [H ~> m or kg m-2]
-
real, dimension( : , : ), allocatable mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::eta_pf instantaneous SSH used in calculating PFu and PFv [H ~> m or kg m-2]
-
real, dimension( : , : ), allocatable mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::uhbt average x-volume or mass flux determined by the barotropic solver [H m2 s-1 ~> m3 s-1 or kg s-1]. uhbt is roughly equal to the vertical sum of uh.
-
real, dimension( : , : ), allocatable mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::vhbt average y-volume or mass flux determined by the barotropic solver [H m2 s-1 ~> m3 s-1 or kg s-1]. vhbt is roughly equal to vertical sum of vh.
-
real, dimension( : , : , : ), allocatable mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::pbce pbce times eta gives the baroclinic pressure anomaly in each layer due to free surface height anomalies [L2 H-1 T-2 ~> m s-2 or m4 kg-1 s-2].
-
real, dimension(:,:), pointer mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::taux_bot => NULL() frictional x-bottom stress from the ocean to the seafloor [Pa]
-
real, dimension(:,:), pointer mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::tauy_bot => NULL() frictional y-bottom stress from the ocean to the seafloor [Pa]
-
type(bt_cont_type), pointer mom_dynamics_split_rk2::mom_dyn_split_rk2_cs::bt_cont => NULL() A structure with elements that describe the effective summed open face areas as a function of barotropic flow.
-
logical
bt_use_layer_fluxes¶ If true, use the summed layered fluxes plus an adjustment due to a changed barotropic velocity in the barotropic continuity equation.
-
logical
split_bottom_stress¶ If true, provide the bottom stress calculated by the vertical viscosity to the barotropic solver.
-
logical
calc_dtbt¶ If true, calculate the barotropic time-step dynamically.
-
real
be¶ A nondimensional number from 0.5 to 1 that controls the backward weighting of the time stepping scheme.
-
real
begw¶ A nondimensional number from 0 to 1 that controls the extent to which the treatment of gravity waves is forward-backward (0) or simulated backward Euler (1). 0 is almost always used.
-
logical
debug¶ If true, write verbose checksums for debugging purposes.
-
logical
debug_obc¶ If true, do debugging calls for open boundary conditions.
-
logical
module_is_initialized= .false.¶ Record whether this mouled has been initialzed.
-
integer
- type
MOM_dynamics_unsplit module control structure.
Unnamed Group
-
integer
id_uh= -1¶ Diagnostic IDs.
-
integer
id_vh= -1¶ Diagnostic IDs.
-
integer
id_pfu= -1¶ Diagnostic IDs.
-
integer
id_pfv= -1¶ Diagnostic IDs.
-
integer
id_cau= -1¶ Diagnostic IDs.
-
integer
id_cav= -1¶ Diagnostic IDs.
-
type(diag_ctrl), pointer mom_dynamics_unsplit::mom_dyn_unsplit_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
type(accel_diag_ptrs), pointer mom_dynamics_unsplit::mom_dyn_unsplit_cs::adp => NULL() A structure pointing to the accelerations in the momentum equations, which can later be used to calculate derived diagnostics like energy budgets.
-
type(cont_diag_ptrs), pointer mom_dynamics_unsplit::mom_dyn_unsplit_cs::cdp => NULL() A structure with pointers to various terms in the continuity equations, which can later be used to calculate derived diagnostics like energy budgets.
-
type(hor_visc_cs), pointer mom_dynamics_unsplit::mom_dyn_unsplit_cs::hor_visc_csp => NULL() A pointer to the horizontal viscosity control structure.
-
type(continuity_cs), pointer mom_dynamics_unsplit::mom_dyn_unsplit_cs::continuity_csp => NULL() A pointer to the continuity control structure.
-
type(coriolisadv_cs), pointer mom_dynamics_unsplit::mom_dyn_unsplit_cs::coriolisadv_csp => NULL() A pointer to the CoriolisAdv control structure.
-
type(pressureforce_cs), pointer mom_dynamics_unsplit::mom_dyn_unsplit_cs::pressureforce_csp => NULL() A pointer to the PressureForce control structure.
-
type(vertvisc_cs), pointer mom_dynamics_unsplit::mom_dyn_unsplit_cs::vertvisc_csp => NULL() A pointer to the vertvisc control structure.
-
type(set_visc_cs), pointer mom_dynamics_unsplit::mom_dyn_unsplit_cs::set_visc_csp => NULL() A pointer to the set_visc control structure.
-
type(tidal_forcing_cs), pointer mom_dynamics_unsplit::mom_dyn_unsplit_cs::tides_csp => NULL() A pointer to the tidal forcing control structure.
-
type(ale_cs), pointer mom_dynamics_unsplit::mom_dyn_unsplit_cs::ale_csp => NULL() A pointer to the ALE control structure.
-
type(ocean_obc_type), pointer mom_dynamics_unsplit::mom_dyn_unsplit_cs::obc => NULL() A pointer to an open boundary.
-
type(update_obc_cs), pointer mom_dynamics_unsplit::mom_dyn_unsplit_cs::update_obc_csp => NULL() A pointer to the update_OBC control structure.
Public Members
-
real, dimension( : , : , : ), allocatable mom_dynamics_unsplit::mom_dyn_unsplit_cs::cau CAu = f*v - u.grad(u) [m s-2].
-
real, dimension( : , : , : ), allocatable mom_dynamics_unsplit::mom_dyn_unsplit_cs::pfu PFu = -dM/dx [m s-2].
-
real, dimension( : , : , : ), allocatable mom_dynamics_unsplit::mom_dyn_unsplit_cs::diffu Zonal acceleration due to convergence of the along-isopycnal stress tensor [m s-1 T-1 ~> mm s-2].
-
real, dimension( : , : , : ), allocatable mom_dynamics_unsplit::mom_dyn_unsplit_cs::cav CAv = -f*u - u.grad(v) [m s-2].
-
real, dimension( : , : , : ), allocatable mom_dynamics_unsplit::mom_dyn_unsplit_cs::pfv PFv = -dM/dy [m s-2].
-
real, dimension( : , : , : ), allocatable mom_dynamics_unsplit::mom_dyn_unsplit_cs::diffv Meridional acceleration due to convergence of the along-isopycnal stress tensor [m s-1 T-1 ~> m s-2].
-
real, dimension(:,:), pointer mom_dynamics_unsplit::mom_dyn_unsplit_cs::taux_bot => NULL() frictional x-bottom stress from the ocean to the seafloor (Pa)
-
real, dimension(:,:), pointer mom_dynamics_unsplit::mom_dyn_unsplit_cs::tauy_bot => NULL() frictional y-bottom stress from the ocean to the seafloor (Pa)
-
logical
debug¶ If true, write verbose checksums for debugging purposes.
-
logical
module_is_initialized= .false.¶ Record whether this mouled has been initialzed.
-
integer
- type
MOM_dynamics_unsplit_RK2 module control structure.
Unnamed Group
-
integer
id_uh= -1¶ Diagnostic IDs.
-
integer
id_vh= -1¶ Diagnostic IDs.
-
integer
id_pfu= -1¶ Diagnostic IDs.
-
integer
id_pfv= -1¶ Diagnostic IDs.
-
integer
id_cau= -1¶ Diagnostic IDs.
-
integer
id_cav= -1¶ Diagnostic IDs.
-
type(diag_ctrl), pointer mom_dynamics_unsplit_rk2::mom_dyn_unsplit_rk2_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
type(accel_diag_ptrs), pointer mom_dynamics_unsplit_rk2::mom_dyn_unsplit_rk2_cs::adp => NULL() A structure pointing to the accelerations in the momentum equations, which can later be used to calculate derived diagnostics like energy budgets.
-
type(cont_diag_ptrs), pointer mom_dynamics_unsplit_rk2::mom_dyn_unsplit_rk2_cs::cdp => NULL() A structure with pointers to various terms in the continuity equations, which can later be used to calculate derived diagnostics like energy budgets.
-
type(hor_visc_cs), pointer mom_dynamics_unsplit_rk2::mom_dyn_unsplit_rk2_cs::hor_visc_csp => NULL() A pointer to the horizontal viscosity control structure.
-
type(continuity_cs), pointer mom_dynamics_unsplit_rk2::mom_dyn_unsplit_rk2_cs::continuity_csp => NULL() A pointer to the continuity control structure.
-
type(coriolisadv_cs), pointer mom_dynamics_unsplit_rk2::mom_dyn_unsplit_rk2_cs::coriolisadv_csp => NULL() A pointer to the CoriolisAdv control structure.
-
type(pressureforce_cs), pointer mom_dynamics_unsplit_rk2::mom_dyn_unsplit_rk2_cs::pressureforce_csp => NULL() A pointer to the PressureForce control structure.
-
type(vertvisc_cs), pointer mom_dynamics_unsplit_rk2::mom_dyn_unsplit_rk2_cs::vertvisc_csp => NULL() A pointer to the vertvisc control structure.
-
type(set_visc_cs), pointer mom_dynamics_unsplit_rk2::mom_dyn_unsplit_rk2_cs::set_visc_csp => NULL() A pointer to the set_visc control structure.
-
type(tidal_forcing_cs), pointer mom_dynamics_unsplit_rk2::mom_dyn_unsplit_rk2_cs::tides_csp => NULL() A pointer to the tidal forcing control structure.
-
type(ale_cs), pointer mom_dynamics_unsplit_rk2::mom_dyn_unsplit_rk2_cs::ale_csp => NULL() A pointer to the ALE control structure.
-
type(ocean_obc_type), pointer mom_dynamics_unsplit_rk2::mom_dyn_unsplit_rk2_cs::obc => NULL() A pointer to an open boundary condition type that specifies whether, where, and what open boundary conditions are used. If no open BCs are used, this pointer stays nullified. Flather OBCs use open boundary_CS as well.
-
type(update_obc_cs), pointer mom_dynamics_unsplit_rk2::mom_dyn_unsplit_rk2_cs::update_obc_csp => NULL() A pointer to the update_OBC control structure.
Public Members
-
real, dimension( : , : , : ), allocatable mom_dynamics_unsplit_rk2::mom_dyn_unsplit_rk2_cs::cau CAu = f*v - u.grad(u) [m s-2].
-
real, dimension( : , : , : ), allocatable mom_dynamics_unsplit_rk2::mom_dyn_unsplit_rk2_cs::pfu PFu = -dM/dx [m s-2].
-
real, dimension( : , : , : ), allocatable mom_dynamics_unsplit_rk2::mom_dyn_unsplit_rk2_cs::diffu Zonal acceleration due to convergence of the along-isopycnal stress tensor [m s-1 T-1 ~> m s-2].
-
real, dimension( : , : , : ), allocatable mom_dynamics_unsplit_rk2::mom_dyn_unsplit_rk2_cs::cav CAv = -f*u - u.grad(v) [m s-2].
-
real, dimension( : , : , : ), allocatable mom_dynamics_unsplit_rk2::mom_dyn_unsplit_rk2_cs::pfv PFv = -dM/dy [m s-2].
-
real, dimension( : , : , : ), allocatable mom_dynamics_unsplit_rk2::mom_dyn_unsplit_rk2_cs::diffv Meridional acceleration due to convergence of the along-isopycnal stress tensor [m s-1 T-1 ~> m s-2].
-
real, dimension(:,:), pointer mom_dynamics_unsplit_rk2::mom_dyn_unsplit_rk2_cs::taux_bot => NULL() frictional x-bottom stress from the ocean to the seafloor (Pa)
-
real, dimension(:,:), pointer mom_dynamics_unsplit_rk2::mom_dyn_unsplit_rk2_cs::tauy_bot => NULL() frictional y-bottom stress from the ocean to the seafloor (Pa)
-
real
be¶ A nondimensional number from 0.5 to 1 that controls the backward weighting of the time stepping scheme.
-
real
begw¶ A nondimensional number from 0 to 1 that controls the extent to which the treatment of gravity waves is forward-backward (0) or simulated backward Euler (1). 0 is almost always used.
-
logical
debug¶ If true, write verbose checksums for debugging purposes.
-
logical
module_is_initialized= .false.¶ Record whether this mouled has been initialzed.
-
integer
-
interface
mom_read_data¶ Read a data field from a file.
Private Functions
-
subroutine
mom_read_data_4d(filename filename, fieldname fieldname, data data, MOM_Domain MOM_Domain, timelevel timelevel, position position, scale scale)¶ This function uses the fms_io function read_data to read a distributed 4-D data field named “fieldname” from file “filename”. Valid values for “position” include CORNER, CENTER, EAST_FACE and NORTH_FACE.
- Parameters
[in] filename: The name of the file to read[in] fieldname: The variable name of the data in the file[inout] data: The 4-dimensional array into which the data should be read[in] mom_domain: The MOM_Domain that describes the decomposition[in] timelevel: The time level in the file to read[in] position: A flag indicating where this data is located[in] scale: A scaling factor that the field is multiplied by before they are returned.
-
subroutine
mom_read_data_3d(filename filename, fieldname fieldname, data data, MOM_Domain MOM_Domain, timelevel timelevel, position position, scale scale)¶ This function uses the fms_io function read_data to read a distributed 3-D data field named “fieldname” from file “filename”. Valid values for “position” include CORNER, CENTER, EAST_FACE and NORTH_FACE.
- Parameters
[in] filename: The name of the file to read[in] fieldname: The variable name of the data in the file[inout] data: The 3-dimensional array into which the data should be read[in] mom_domain: The MOM_Domain that describes the decomposition[in] timelevel: The time level in the file to read[in] position: A flag indicating where this data is located[in] scale: A scaling factor that the field is multiplied by before they are returned.
-
subroutine
mom_read_data_2d(filename filename, fieldname fieldname, data data, MOM_Domain MOM_Domain, timelevel timelevel, position position, scale scale)¶ This function uses the fms_io function read_data to read a distributed 2-D data field named “fieldname” from file “filename”. Valid values for “position” include CORNER, CENTER, EAST_FACE and NORTH_FACE.
- Parameters
[in] filename: The name of the file to read[in] fieldname: The variable name of the data in the file[inout] data: The 2-dimensional array into which the data should be read[in] mom_domain: The MOM_Domain that describes the decomposition[in] timelevel: The time level in the file to read[in] position: A flag indicating where this data is located[in] scale: A scaling factor that the field is multiplied by before they are returned.
-
subroutine
mom_read_data_1d(filename filename, fieldname fieldname, data data, timelevel timelevel, scale scale)¶ This function uses the fms_io function read_data to read 1-D data field named “fieldname” from file “filename”.
- Parameters
[in] filename: The name of the file to read[in] fieldname: The variable name of the data in the file[inout] data: The 1-dimensional array into which the data[in] timelevel: The time level in the file to read[in] scale: A scaling factor that the field is multiplied by before they are returned.
-
subroutine
-
interface
mom_read_vector¶ Read a pair of data fields representing the two components of a vector from a file.
Private Functions
-
subroutine
mom_read_vector_3d(filename filename, u_fieldname u_fieldname, v_fieldname v_fieldname, u_data u_data, v_data v_data, MOM_Domain MOM_Domain, timelevel timelevel, stagger stagger, scalar_pair scalar_pair, scale scale)¶ This function uses the fms_io function read_data to read a pair of distributed 3-D data fields with names given by “[uv]_fieldname” from file “filename”. Valid values for “stagger” include CGRID_NE, BGRID_NE, and AGRID.
- Parameters
[in] filename: The name of the file to read[in] u_fieldname: The variable name of the u data in the file[in] v_fieldname: The variable name of the v data in the file[inout] u_data: The 3 dimensional array into which the u-component of the data should be read[inout] v_data: The 3 dimensional array into which the v-component of the data should be read[in] mom_domain: The MOM_Domain that describes the decomposition[in] timelevel: The time level in the file to read[in] stagger: A flag indicating where this vector is discretized[in] scalar_pair: If true, a pair of scalars are to be read.cretized[in] scale: A scaling factor that the fields are multiplied by before they are returned.
-
subroutine
mom_read_vector_2d(filename filename, u_fieldname u_fieldname, v_fieldname v_fieldname, u_data u_data, v_data v_data, MOM_Domain MOM_Domain, timelevel timelevel, stagger stagger, scalar_pair scalar_pair, scale scale)¶ This function uses the fms_io function read_data to read a pair of distributed 2-D data fields with names given by “[uv]_fieldname” from file “filename”. Valid values for “stagger” include CGRID_NE, BGRID_NE, and AGRID.
- Parameters
[in] filename: The name of the file to read[in] u_fieldname: The variable name of the u data in the file[in] v_fieldname: The variable name of the v data in the file[inout] u_data: The 2 dimensional array into which the u-component of the data should be read[inout] v_data: The 2 dimensional array into which the v-component of the data should be read[in] mom_domain: The MOM_Domain that describes the decomposition[in] timelevel: The time level in the file to read[in] stagger: A flag indicating where this vector is discretized[in] scalar_pair: If true, a pair of scalars are to be read.cretized[in] scale: A scaling factor that the fields are multiplied by before they are returned.
-
subroutine
- type
A restart registry and the control structure for restarts.
Unnamed Group
-
type(p0d), dimension(:), pointer mom_restart::mom_restart_cs::var_ptr0d => NULL() Pointers to the fields that have been registered for restarts.
-
type(p1d), dimension(:), pointer mom_restart::mom_restart_cs::var_ptr1d => NULL() Pointers to the fields that have been registered for restarts.
-
type(p2d), dimension(:), pointer mom_restart::mom_restart_cs::var_ptr2d => NULL() Pointers to the fields that have been registered for restarts.
-
type(p3d), dimension(:), pointer mom_restart::mom_restart_cs::var_ptr3d => NULL() Pointers to the fields that have been registered for restarts.
-
type(p4d), dimension(:), pointer mom_restart::mom_restart_cs::var_ptr4d => NULL() Pointers to the fields that have been registered for restarts.
-
integer
max_fields¶ The maximum number of restart fields.
Public Members
-
logical
restart¶ restart is set to .true. if the run has been started from a full restart file. Otherwise some fields must be initialized approximately.
-
integer
novars= 0¶ The number of restart fields that have been registered.
-
integer
num_obsolete_vars= 0¶ The number of obsolete restart fields that have been registered.
-
logical
parallel_restartfiles¶ If true, each PE writes its own restart file, otherwise they are combined internally.
-
logical
large_file_support¶ If true, NetCDF 3.6 or later is being used and large-file-support is enabled.
-
logical
new_run¶ If true, the input filenames and restart file existence will result in a new run that is not initialized from restart files.
-
logical
new_run_set= .false.¶ If true, new_run has been determined for this restart_CS.
-
logical
checksum_required¶ If true, require the restart checksums to match and error out otherwise. Users may want to avoid this comparison if for example the restarts are made from a run with a different mask_table than the current run, in which case the checksums will not match and cause crash.
-
character(len=240) mom_restart::mom_restart_cs::restartfile The name or name root for MOM restart files.
-
type(field_restart), dimension(:), pointer mom_restart::mom_restart_cs::restart_field => NULL() An array of descriptions of the registered fields.
-
type(obsolete_restart), dimension(:), pointer mom_restart::mom_restart_cs::restart_obsolete => NULL() An array of obsolete restart fields.
-
-
interface
mom_state_chksum¶ Write out checksums of the MOM6 state variables.
Private Functions
-
subroutine
mom_state_chksum_5arg(mesg mesg, u u, v v, h h, uh uh, vh vh, G G, GV GV, haloshift haloshift, symmetric symmetric)¶ Write out chksums for the model’s basic state variables, including transports.
- Parameters
[in] mesg: A message that appears on the chksum lines.[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] u: The zonal velocity [m s-1].[in] v: The meridional velocity [m s-1].[in] h: Layer thicknesses [H ~> m or kg m-2].[in] uh: Volume flux through zonal faces = u*h*dy[in] vh: Volume flux through meridional faces = v*h*dx[in] haloshift: The width of halos to check (default 0).[in] symmetric: If true, do checksums on the fully symmetric computationoal domain.
-
subroutine
mom_state_chksum_3arg(mesg mesg, u u, v v, h h, G G, GV GV, haloshift haloshift, symmetric symmetric)¶ Write out chksums for the model’s basic state variables.
- Parameters
[in] mesg: A message that appears on the chksum lines.[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] u: Zonal velocity [m s-1].[in] v: Meridional velocity [m s-1].[in] h: Layer thicknesses [H ~> m or kg m-2].[in] haloshift: The width of halos to check (default 0).[in] symmetric: If true, do checksums on the fully symmetric computationoal domain.
-
subroutine
- type
The control structure for this module.
Public Members
-
integer
nterm¶ Number of terms in polynomial (deg+1)
-
integer
max_iter¶ Maximum number of iterations.
-
real
drho_tol¶ Tolerance criterion for difference in density [kg m-3].
-
real
xtol¶ Criterion for how much position changes [nondim].
-
real
ref_pres¶ Determines whether a constant reference pressure is used everywhere or locally referenced density is done. ref_pres <-1 is the latter, ref_pres >= 0. otherwise.
-
logical
force_brent= .false.¶ Use Brent’s method instead of Newton even when second derivatives are available.
-
logical
debug¶ If true, write verbose debugging messages and checksusm.
-
type(eos_type), pointer mom_neutral_diffusion_aux::ndiff_aux_cs_type::eos Pointer to equation of state used in the model.
-
integer
- type
The control structure for the MOM_neutral_diffusion module.
Public Members
-
integer
nkp1¶ Number of interfaces for a column = nk + 1.
-
integer
nsurf¶ Number of neutral surfaces.
-
integer
deg= 2¶ Degree of polynomial used for reconstructions.
-
logical
continuous_reconstruction= .true.¶ True if using continuous PPM reconstruction at interfaces.
-
logical
refine_position= .false.¶ If true, iterate to refine the corresponding positions in neighboring columns.
-
logical
debug= .false.¶ If true, write verbose debugging messages.
-
integer
max_iter¶ Maximum number of iterations if refine_position is defined.
-
real
tolerance¶ Convergence criterion representing difference from true neutrality.
-
real
ref_pres¶ Reference pressure, negative if using locally referenced neutral density.
-
real, dimension(:,:,:), allocatable mom_neutral_diffusion::neutral_diffusion_cs::upol Non-dimensional position with left layer uKoL-1, u-point.
-
real, dimension(:,:,:), allocatable mom_neutral_diffusion::neutral_diffusion_cs::upor Non-dimensional position with right layer uKoR-1, u-point.
-
integer, dimension(:,:,:), allocatable mom_neutral_diffusion::neutral_diffusion_cs::ukol Index of left interface corresponding to neutral surface, at a u-point.
-
integer, dimension(:,:,:), allocatable mom_neutral_diffusion::neutral_diffusion_cs::ukor Index of right interface corresponding to neutral surface, at a u-point.
-
real, dimension(:,:,:), allocatable mom_neutral_diffusion::neutral_diffusion_cs::uheff Effective thickness at u-point [H ~> m or kg m-2].
-
real, dimension(:,:,:), allocatable mom_neutral_diffusion::neutral_diffusion_cs::vpol Non-dimensional position with left layer uKoL-1, v-point.
-
real, dimension(:,:,:), allocatable mom_neutral_diffusion::neutral_diffusion_cs::vpor Non-dimensional position with right layer uKoR-1, v-point.
-
integer, dimension(:,:,:), allocatable mom_neutral_diffusion::neutral_diffusion_cs::vkol Index of left interface corresponding to neutral surface, at a v-point.
-
integer, dimension(:,:,:), allocatable mom_neutral_diffusion::neutral_diffusion_cs::vkor Index of right interface corresponding to neutral surface, at a v-point.
-
real, dimension(:,:,:), allocatable mom_neutral_diffusion::neutral_diffusion_cs::vheff Effective thickness at v-point [H ~> m or kg m-2].
-
real, dimension(:,:,:,:), allocatable mom_neutral_diffusion::neutral_diffusion_cs::ppoly_coeffs_t Polynomial coefficients for temperature.
-
real, dimension(:,:,:,:), allocatable mom_neutral_diffusion::neutral_diffusion_cs::ppoly_coeffs_s Polynomial coefficients for salinity.
-
real, dimension(:,:,:), allocatable mom_neutral_diffusion::neutral_diffusion_cs::drdt dRho/dT [kg m-3 degC-1] at interfaces
-
real, dimension(:,:,:), allocatable mom_neutral_diffusion::neutral_diffusion_cs::drds dRho/dS [kg m-3 ppt-1] at interfaces
-
real, dimension(:,:,:), allocatable mom_neutral_diffusion::neutral_diffusion_cs::tint Interface T [degC].
-
real, dimension(:,:,:), allocatable mom_neutral_diffusion::neutral_diffusion_cs::sint Interface S [ppt].
-
real, dimension(:,:,:), allocatable mom_neutral_diffusion::neutral_diffusion_cs::pint Interface pressure [Pa].
-
real, dimension(:,:,:,:), allocatable mom_neutral_diffusion::neutral_diffusion_cs::t_i Top edge reconstruction of temperature [degC].
-
real, dimension(:,:,:,:), allocatable mom_neutral_diffusion::neutral_diffusion_cs::s_i Top edge reconstruction of salinity [ppt].
-
real, dimension(:,:,:,:), allocatable mom_neutral_diffusion::neutral_diffusion_cs::drdt_i dRho/dT [kg m-3 degC-1] at top edge
-
real, dimension(:,:,:,:), allocatable mom_neutral_diffusion::neutral_diffusion_cs::drds_i dRho/dS [kg m-3 ppt-1] at top edge
-
integer, dimension(:,:), allocatable mom_neutral_diffusion::neutral_diffusion_cs::ns Number of interfacs in a column.
-
logical, dimension(:,:,:), allocatable mom_neutral_diffusion::neutral_diffusion_cs::stable_cell True if the cell is stably stratified wrt to the next cell.
-
type(diag_ctrl), pointer mom_neutral_diffusion::neutral_diffusion_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
integer
id_uheff_2d= -1¶ Diagnostic IDs.
-
integer
id_vheff_2d= -1¶ Diagnostic IDs.
-
real
c_p¶ heat capacity of seawater (J kg-1 K-1)
-
type(eos_type), pointer mom_neutral_diffusion::neutral_diffusion_cs::eos Equation of state parameters.
-
type(remapping_cs) mom_neutral_diffusion::neutral_diffusion_cs::remap_cs Remapping control structure used to create sublayers.
-
type(ndiff_aux_cs_type), pointer mom_neutral_diffusion::neutral_diffusion_cs::ndiff_aux_cs Store parameters for iteratively finding neutral surface.
-
integer
- type
This control structure should be used to store any run-time variables associated with the Neverland forcing.
It can be readily modified for a specific case, and because it is private there will be no changes needed in other code (although they will have to be recompiled).
Public Members
-
logical
use_temperature¶ If true, use temperature and salinity.
-
logical
restorebuoy¶ If true, use restoring surface buoyancy forcing.
-
real
rho0¶ The density used in the Boussinesq approximation [kg m-3].
-
real
g_earth¶ The gravitational acceleration [L2 Z-1 T-2 ~> m s-2].
-
real
flux_const¶ The restoring rate at the surface [m s-1].
-
real, dimension(:,:), pointer neverland_surface_forcing::neverland_surface_forcing_cs::buoy_restore => NULL() The pattern to restore buoyancy to.
-
character(len=200) neverland_surface_forcing::neverland_surface_forcing_cs::inputdir The directory where NetCDF input files are.
-
type(diag_ctrl), pointer neverland_surface_forcing::neverland_surface_forcing_cs::diag A structure that is used to regulate the timing of diagnostic output.
-
logical
first_call= .true.¶ True until Neverland_buoyancy_forcing has been called.
-
logical
- type
Type to carry basic OBC information needed for updating values.
Public Members
-
integer
nobc= 0¶ number of registered open boundary types.
-
type(obc_struct_type), dimension( 50 ) mom_open_boundary::obc_registry_type::ob array of registered boundary types.
-
logical
locked= .false.¶ New OBC types may be registered if locked=.false. When locked=.true.,no more boundaries can be registered.
-
integer
- type
Open boundary segment data from files (mostly).
Public Members
-
integer
fid¶ handle from FMS associated with segment data on disk
-
integer
fid_dz¶ handle from FMS associated with segment thicknesses on disk
-
character(len=8) mom_open_boundary::obc_segment_data_type::name a name identifier for the segment data
-
real, dimension(:,:,:), pointer mom_open_boundary::obc_segment_data_type::buffer_src =>NULL() buffer for segment data located at cell faces and on the original vertical grid
-
integer
nk_src¶ Number of vertical levels in the source data.
-
real, dimension(:,:,:), pointer mom_open_boundary::obc_segment_data_type::dz_src =>NULL() vertical grid cell spacing of the incoming segment data [m]
-
real, dimension(:,:,:), pointer mom_open_boundary::obc_segment_data_type::buffer_dst =>NULL() buffer src data remapped to the target vertical grid
-
real, dimension(:,:), pointer mom_open_boundary::obc_segment_data_type::bt_vel =>NULL() barotropic velocity [m s-1]
-
real
value¶ constant value if fid is equal to -1
-
integer
- type
Tracer segment data structure, for putting into an array of objects, not all the same shape.
Tracer on OBC segment data structure, for putting into a segment tracer registry.
Public Members
-
real, dimension(:,:,:), pointer mom_open_boundary::obc_segment_tracer_type::t => NULL() tracer concentration array
-
real
obc_inflow_conc= 0.0¶ tracer concentration for generic inflows
-
character(len=32) mom_open_boundary::obc_segment_tracer_type::name tracer name used for error messages
-
type(tracer_type), pointer mom_open_boundary::obc_segment_tracer_type::tr => NULL() metadata describing the tracer
-
real, dimension(:,:,:), pointer mom_open_boundary::obc_segment_tracer_type::tres => NULL() tracer reservoir array
-
logical
is_initialized¶ reservoir values have been set when True
-
- type
Open boundary segment data structure.
Public Members
-
logical
flather¶ If true, applies Flather + Chapman radiation of barotropic gravity waves.
-
logical
radiation¶ If true, 1D Orlanksi radiation boundary conditions are applied. If False, a gradient condition is applied.
-
logical
radiation_tan¶ If true, 1D Orlanksi radiation boundary conditions are applied to tangential flows.
-
logical
radiation_grad¶ If true, 1D Orlanksi radiation boundary conditions are applied to dudv and dvdx.
-
logical
oblique¶ Oblique waves supported at radiation boundary.
-
logical
oblique_tan¶ If true, 2D radiation boundary conditions are applied to tangential flows.
-
logical
oblique_grad¶ If true, 2D radiation boundary conditions are applied to dudv and dvdx.
-
logical
nudged¶ Optional supplement to radiation boundary.
-
logical
nudged_tan¶ Optional supplement to nudge tangential velocity.
-
logical
nudged_grad¶ Optional supplement to nudge normal gradient of tangential velocity.
-
logical
specified¶ Boundary normal velocity fixed to external value.
-
logical
specified_tan¶ Boundary tangential velocity fixed to external value.
-
logical
open¶ Boundary is open for continuity solver.
-
logical
gradient¶ Zero gradient at boundary.
-
logical
values_needed¶ Whether or not external OBC fields are needed.
-
integer
direction¶ Boundary faces one of the four directions.
-
logical
is_n_or_s¶ True is the OB is facing North or South and exists on this PE.
-
logical
is_e_or_w¶ True is the OB is facing East or West and exists on this PE.
-
type(obc_segment_data_type), dimension(:), pointer mom_open_boundary::obc_segment_type::field =>NULL() OBC data.
-
integer
num_fields¶ number of OBC data fields (e.g. u_normal,u_parallel and eta for Flather)
-
character(len=32), dimension(:), pointer mom_open_boundary::obc_segment_type::field_names =>NULL() field names for this segment
-
integer
is_obc¶ i-indices of boundary segment.
-
integer
ie_obc¶ i-indices of boundary segment.
-
integer
js_obc¶ j-indices of boundary segment.
-
integer
je_obc¶ j-indices of boundary segment.
-
real
velocity_nudging_timescale_in¶ Nudging timescale on inflow [s].
-
real
velocity_nudging_timescale_out¶ Nudging timescale on outflow [s].
-
logical
on_pe¶ true if segment is located in the computational domain
-
logical
temp_segment_data_exists¶ true if temperature data arrays are present
-
logical
salt_segment_data_exists¶ true if salinity data arrays are present
-
real, dimension(:,:), pointer mom_open_boundary::obc_segment_type::cg =>NULL() The external gravity wave speed [m s-1] at OBC-points.
-
real, dimension(:,:), pointer mom_open_boundary::obc_segment_type::htot =>NULL() The total column thickness [m] at OBC-points.
-
real, dimension(:,:,:), pointer mom_open_boundary::obc_segment_type::h =>NULL() The cell thickness [m] at OBC-points.
-
real, dimension(:,:,:), pointer mom_open_boundary::obc_segment_type::normal_vel =>NULL() The layer velocity normal to the OB segment [m s-1].
-
real, dimension(:,:,:), pointer mom_open_boundary::obc_segment_type::tangential_vel =>NULL() The layer velocity tangential to the OB segment [m s-1].
-
real, dimension(:,:,:), pointer mom_open_boundary::obc_segment_type::tangential_grad =>NULL() The gradient of the velocity tangential to the OB segment [m s-1].
-
real, dimension(:,:,:), pointer mom_open_boundary::obc_segment_type::normal_trans =>NULL() The layer transport normal to the OB segment [m3 s-1].
-
real, dimension(:,:), pointer mom_open_boundary::obc_segment_type::normal_vel_bt =>NULL() The barotropic velocity normal to the OB segment [m s-1].
-
real, dimension(:,:), pointer mom_open_boundary::obc_segment_type::eta =>NULL() The sea-surface elevation along the segment [m].
-
real, dimension(:,:,:), pointer mom_open_boundary::obc_segment_type::grad_normal =>NULL() The gradient of the normal flow along the segment [s-1].
-
real, dimension(:,:,:), pointer mom_open_boundary::obc_segment_type::grad_tan =>NULL() The gradient of the tangential flow along the segment [s-1].
-
real, dimension(:,:,:), pointer mom_open_boundary::obc_segment_type::grad_gradient =>NULL() The gradient of the gradient of tangential flow along the segment [m-1 s-1].
-
real, dimension(:,:,:), pointer mom_open_boundary::obc_segment_type::rx_normal =>NULL() The rx_old_u value for radiation coeff for normal velocity.
-
real, dimension(:,:,:), pointer mom_open_boundary::obc_segment_type::ry_normal =>NULL() The tangential value for radiation coeff for normal velocity.
-
real, dimension(:,:,:), pointer mom_open_boundary::obc_segment_type::cff_normal =>NULL() The denominator for oblique radiation for normal velocity.
-
real, dimension(:,:,:), pointer mom_open_boundary::obc_segment_type::nudged_normal_vel =>NULL() The layer velocity normal to the OB segment that values should be nudged towards [m s-1].
-
real, dimension(:,:,:), pointer mom_open_boundary::obc_segment_type::nudged_tangential_vel =>NULL() The layer velocity tangential to the OB segment that values should be nudged towards [m s-1].
-
real, dimension(:,:,:), pointer mom_open_boundary::obc_segment_type::nudged_tangential_grad =>NULL() The layer dvdx or dudy towards which nudging can occur [s-1].
-
type(segment_tracer_registry_type), pointer mom_open_boundary::obc_segment_type::tr_reg => NULL() A pointer to the tracer registry for the segment.
-
type(hor_index_type) mom_open_boundary::obc_segment_type::hi Horizontal index ranges.
-
real
tr_invlscale3_out¶ An effective inverse length scale cubed [m-3].
-
real
tr_invlscale3_in¶ for restoring the tracer concentration in a ficticious reservior towards interior values when flow is exiting the domain, or towards an externally imposed value when flow is entering
-
logical
- type
Type to carry something (what] for the OBC registry.
Public Members
-
character(len=32) mom_open_boundary::obc_struct_type::name OBC name used for error messages.
-
- type
A structure to store information about restart fields that are no longer used.
Private Members
-
character(len=32) mom_restart::obsolete_restart::field_name Name of restart field that is no longer in use.
-
character(len=32) mom_restart::obsolete_restart::replacement_name Name of replacement restart field, if applicable.
-
- type
Ocean grid type. See mom_grid for details.
Public Members
-
type(mom_domain_type), pointer mom_grid::ocean_grid_type::domain => NULL() Ocean model domain.
-
type(mom_domain_type), pointer mom_grid::ocean_grid_type::domain_aux => NULL() A non-symmetric auxiliary domain type.
-
type(hor_index_type) mom_grid::ocean_grid_type::hi Horizontal index ranges.
-
type(hor_index_type) mom_grid::ocean_grid_type::hid2 Horizontal index ranges for level-2-downsampling.
-
integer
isc¶ The start i-index of cell centers within the computational domain.
-
integer
iec¶ The end i-index of cell centers within the computational domain.
-
integer
jsc¶ The start j-index of cell centers within the computational domain.
-
integer
jec¶ The end j-index of cell centers within the computational domain.
-
integer
isd¶ The start i-index of cell centers within the data domain.
-
integer
ied¶ The end i-index of cell centers within the data domain.
-
integer
jsd¶ The start j-index of cell centers within the data domain.
-
integer
jed¶ The end j-index of cell centers within the data domain.
-
integer
isg¶ The start i-index of cell centers within the global domain.
-
integer
ieg¶ The end i-index of cell centers within the global domain.
-
integer
jsg¶ The start j-index of cell centers within the global domain.
-
integer
jeg¶ The end j-index of cell centers within the global domain.
-
integer
iscb¶ The start i-index of cell vertices within the computational domain.
-
integer
iecb¶ The end i-index of cell vertices within the computational domain.
-
integer
jscb¶ The start j-index of cell vertices within the computational domain.
-
integer
jecb¶ The end j-index of cell vertices within the computational domain.
-
integer
isdb¶ The start i-index of cell vertices within the data domain.
-
integer
iedb¶ The end i-index of cell vertices within the data domain.
-
integer
jsdb¶ The start j-index of cell vertices within the data domain.
-
integer
jedb¶ The end j-index of cell vertices within the data domain.
-
integer
isgb¶ The start i-index of cell vertices within the global domain.
-
integer
iegb¶ The end i-index of cell vertices within the global domain.
-
integer
jsgb¶ The start j-index of cell vertices within the global domain.
-
integer
jegb¶ The end j-index of cell vertices within the global domain.
-
integer
isd_global¶ The value of isd in the global index space (decompoistion invariant).
-
integer
jsd_global¶ The value of isd in the global index space (decompoistion invariant).
-
integer
idg_offset¶ The offset between the corresponding global and local i-indices.
-
integer
jdg_offset¶ The offset between the corresponding global and local j-indices.
-
integer
ke¶ The number of layers in the vertical.
-
logical
symmetric¶ True if symmetric memory is used.
-
logical
nonblocking_updates¶ If true, non-blocking halo updates are allowed. The default is .false. (for now).
-
integer
first_direction¶ An integer that indicates which direction is to be updated first in directionally split parts of the calculation. This can be altered during the course of the run via calls to set_first_direction.
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::mask2dt 0 for land points and 1 for ocean points on the h-grid. Nd.
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::geolatt The geographic latitude at q points in degrees of latitude or m.
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::geolont The geographic longitude at q points in degrees of longitude or m.
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::dxt dxT is delta x at h points [m].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::idxt 1/dxT [m-1].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::dyt dyT is delta y at h points [m].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::idyt IdyT is 1/dyT [m-1].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::areat The area of an h-cell [m2].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::iareat 1/areaT [m-2].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::sin_rot The sine of the angular rotation between the local model grid’s northward.
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::cos_rot The cosine of the angular rotation between the local model grid’s northward.
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::mask2dcu 0 for boundary points and 1 for ocean points on the u grid. Nondim.
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::geolatcu The geographic latitude at u points in degrees of latitude or m.
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::geoloncu The geographic longitude at u points in degrees of longitude or m.
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::dxcu dxCu is delta x at u points [m].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::idxcu 1/dxCu [m-1].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::dycu dyCu is delta y at u points [m].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::idycu 1/dyCu [m-1].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::dy_cu The unblocked lengths of the u-faces of the h-cell [m].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::iareacu The masked inverse areas of u-grid cells [m2].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::areacu The areas of the u-grid cells [m2].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::mask2dcv 0 for boundary points and 1 for ocean points on the v grid. Nondim.
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::geolatcv The geographic latitude at v points in degrees of latitude or m.
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::geoloncv The geographic longitude at v points in degrees of longitude or m.
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::dxcv dxCv is delta x at v points [m].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::idxcv 1/dxCv [m-1].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::dycv dyCv is delta y at v points [m].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::idycv 1/dyCv [m-1].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::dx_cv The unblocked lengths of the v-faces of the h-cell [m].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::iareacv The masked inverse areas of v-grid cells [m2].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::areacv The areas of the v-grid cells [m2].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::mask2dbu 0 for boundary points and 1 for ocean points on the q grid. Nondim.
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::geolatbu The geographic latitude at q points in degrees of latitude or m.
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::geolonbu The geographic longitude at q points in degrees of longitude or m.
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::dxbu dxBu is delta x at q points [m].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::idxbu 1/dxBu [m-1].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::dybu dyBu is delta y at q points [m].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::idybu 1/dyBu [m-1].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::areabu areaBu is the area of a q-cell [m2]
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::iareabu IareaBu = 1/areaBu [m-2].
-
real, dimension(:), pointer mom_grid::ocean_grid_type::gridlatt => NULL() The latitude of T points for the purpose of labeling the output axes.
-
real, dimension(:), pointer mom_grid::ocean_grid_type::gridlatb => NULL() The latitude of B points for the purpose of labeling the output axes.
-
real, dimension(:), pointer mom_grid::ocean_grid_type::gridlont => NULL() The longitude of T points for the purpose of labeling the output axes.
-
real, dimension(:), pointer mom_grid::ocean_grid_type::gridlonb => NULL() The longitude of B points for the purpose of labeling the output axes.
-
character(len=40) mom_grid::ocean_grid_type::x_axis_units The units that are used in labeling the x coordinate axes.
-
character(len=40) mom_grid::ocean_grid_type::y_axis_units The units that are used in labeling the y coordinate axes.
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::bathyt Ocean bottom depth at tracer points, in depth units [Z ~> m].
-
logical
bathymetry_at_vel¶ If true, there are separate values for the basin depths at velocity points. Otherwise the effects of of topography are entirely determined from thickness points.
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::dblock_u Topographic depths at u-points at which the flow is blocked [Z ~> m].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::dopen_u Topographic depths at u-points at which the flow is open at width dy_Cu [Z ~> m].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::dblock_v Topographic depths at v-points at which the flow is blocked [Z ~> m].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::dopen_v Topographic depths at v-points at which the flow is open at width dx_Cv [Z ~> m].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::coriolisbu The Coriolis parameter at corner points [T-1 ~> s-1].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::df_dx Derivative d/dx f (Coriolis parameter) at h-points [T-1 m-1 ~> s-1 m-1].
-
real, dimension( : , : ), allocatable mom_grid::ocean_grid_type::df_dy Derivative d/dy f (Coriolis parameter) at h-points [T-1 m-1 ~> s-1 m-1].
-
real
g_earth¶ The gravitational acceleration [m2 Z-1 s-2 ~> m s-2].
-
real
areat_global¶ Global sum of h-cell area [m2].
-
real
iareat_global¶ Global sum of inverse h-cell area (1/areaT_global) [m2].
-
integer
nblocks¶ The number of sub-PE blocks on this PE.
-
type(hor_index_type), dimension(:), pointer mom_grid::ocean_grid_type::block => NULL() Index ranges for each block.
-
real
south_lat¶ The latitude (or y-coordinate) of the first v-line.
-
real
west_lon¶ The longitude (or x-coordinate) of the first u-line.
-
real
len_lat= 0.¶ The latitudinal (or y-coord) extent of physical domain.
-
real
len_lon= 0.¶ The longitudinal (or x-coord) extent of physical domain.
-
real
rad_earth= 6.378e6¶ The radius of the planet [m].
-
real
max_depth¶ The maximum depth of the ocean in depth units [Z ~> m].
-
- type
Pointers to all of the prognostic variables allocated in MOM_variables.F90 and MOM.F90.
It is useful for sending these variables for diagnostics, and in preparation for ensembles later on. All variables have the same names as the local (public) variables they refer to in MOM.F90.
Public Members
-
real, dimension(:,:,:), pointer mom_variables::ocean_internal_state::t => NULL() Pointer to the temperature state variable [degC].
-
real, dimension(:,:,:), pointer mom_variables::ocean_internal_state::s => NULL() Pointer to the salinity state variable [ppt ~> PSU or g/kg].
-
real, dimension(:,:,:), pointer mom_variables::ocean_internal_state::u => NULL() Pointer to the zonal velocity [m s-1].
-
real, dimension(:,:,:), pointer mom_variables::ocean_internal_state::v => NULL() Pointer to the meridional velocity [m s-1].
-
real, dimension(:,:,:), pointer mom_variables::ocean_internal_state::h => NULL() Pointer to the layer thicknesses [H ~> m or kg m-2].
-
real, dimension(:,:,:), pointer mom_variables::ocean_internal_state::uh => NULL() Pointer to zonal transports [H m2 s-1 ~> m3 s-1 or kg s-1].
-
real, dimension(:,:,:), pointer mom_variables::ocean_internal_state::vh => NULL() Pointer to meridional transports [H m2 s-1 ~> m3 s-1 or kg s-1].
-
real, dimension(:,:,:), pointer mom_variables::ocean_internal_state::cau => NULL() Pointer to the zonal Coriolis and Advective acceleration [m s-2].
-
real, dimension(:,:,:), pointer mom_variables::ocean_internal_state::cav => NULL() Pointer to the meridional Coriolis and Advective acceleration [m s-2].
-
real, dimension(:,:,:), pointer mom_variables::ocean_internal_state::pfu => NULL() Pointer to the zonal Pressure force acceleration [m s-2].
-
real, dimension(:,:,:), pointer mom_variables::ocean_internal_state::pfv => NULL() Pointer to the meridional Pressure force acceleration [m s-2].
-
real, dimension(:,:,:), pointer mom_variables::ocean_internal_state::diffu => NULL() Pointer to the zonal acceleration due to lateral viscosity [m s-1 T-1 ~> m s-2].
-
real, dimension(:,:,:), pointer mom_variables::ocean_internal_state::diffv => NULL() Pointer to the meridional acceleration due to lateral viscosity [m s-1 T-1 ~> m s-2].
-
real, dimension(:,:,:), pointer mom_variables::ocean_internal_state::pbce => NULL() Pointer to the baroclinic pressure force dependency on free surface movement.
-
real, dimension(:,:,:), pointer mom_variables::ocean_internal_state::u_accel_bt => NULL() Pointer to the zonal barotropic-solver acceleration [m s-2].
-
real, dimension(:,:,:), pointer mom_variables::ocean_internal_state::v_accel_bt => NULL() Pointer to the meridional barotropic-solver acceleration [m s-2].
-
real, dimension(:,:,:), pointer mom_variables::ocean_internal_state::u_av => NULL() Pointer to zonal velocity averaged over the timestep [m s-1].
-
real, dimension(:,:,:), pointer mom_variables::ocean_internal_state::v_av => NULL() Pointer to meridional velocity averaged over the timestep [m s-1].
-
real, dimension(:,:,:), pointer mom_variables::ocean_internal_state::u_prev => NULL() Pointer to zonal velocity at the end of the last timestep [m s-1].
-
real, dimension(:,:,:), pointer mom_variables::ocean_internal_state::v_prev => NULL() Pointer to meridional velocity at the end of the last timestep [m s-1].
-
-
interface
ocean_model_data_get¶ This interface extracts a named scalar field or array from the ocean surface or public type.
Private Functions
-
subroutine
ocean_model_data1d_get(OS OS, Ocean Ocean, name name, value value)¶ This subroutine extracts a named scalar field from the ocean surface or public type.
- Parameters
os: A pointer to the structure containing the internal ocean state (intent in).[in] ocean: A structure containing various publicly visible ocean surface fields.[in] name: The name of the field to extract[out] value: The value of the named field
-
subroutine
ocean_model_data2d_get(OS OS, Ocean Ocean, name name, array2D array2D, isc isc, jsc jsc)¶ This subroutine extracts a named 2-D field from the ocean surface or public type.
- Parameters
os: A pointer to the structure containing the internal ocean state (intent in).[in] ocean: A structure containing various publicly visible ocean surface fields.[in] name: The name of the field to extract[out] array2d: The values of the named field, it must cover only the computational domain[in] isc: The starting i-index of array2D[in] jsc: The starting j-index of array2D
-
subroutine
- type
Open-boundary data.
Public Members
-
integer
number_of_segments= 0¶ The number of open-boundary segments.
-
integer
ke= 0¶ The number of model layers.
-
logical
open_u_bcs_exist_globally= .false.¶ True if any zonal velocity points in the global domain use open BCs.
-
logical
open_v_bcs_exist_globally= .false.¶ True if any meridional velocity points in the global domain use open BCs.
-
logical
flather_u_bcs_exist_globally= .false.¶ True if any zonal velocity points in the global domain use Flather BCs.
-
logical
flather_v_bcs_exist_globally= .false.¶ True if any meridional velocity points in the global domain use Flather BCs.
-
logical
oblique_bcs_exist_globally= .false.¶ True if any velocity points in the global domain use oblique BCs.
-
logical
nudged_u_bcs_exist_globally= .false.¶ True if any velocity points in the global domain use nudged BCs.
-
logical
nudged_v_bcs_exist_globally= .false.¶ True if any velocity points in the global domain use nudged BCs.
-
logical
specified_u_bcs_exist_globally= .false.¶ True if any zonal velocity points in the global domain use specified BCs.
-
logical
specified_v_bcs_exist_globally= .false.¶ True if any meridional velocity points in the global domain use specified BCs.
-
logical
radiation_bcs_exist_globally= .false.¶ True if radiations BCs are in use anywhere.
-
logical
user_bcs_set_globally= .false.¶ True if any OBC_USER_CONFIG is set for input from user directory.
-
logical
update_obc= .false.¶ Is OBC data time-dependent.
-
logical
needs_io_for_data= .false.¶ Is any i/o needed for OBCs.
-
logical
zero_vorticity= .false.¶ If True, sets relative vorticity to zero on open boundaries.
-
logical
freeslip_vorticity= .false.¶ If True, sets normal gradient of tangential velocity to zero in the relative vorticity on open boundaries.
-
logical
computed_vorticity= .false.¶ If True, uses external data for tangential velocity in the relative vorticity on open boundaries.
-
logical
specified_vorticity= .false.¶ If True, uses external data for tangential velocity gradients in the relative vorticity on open boundaries.
-
logical
zero_strain= .false.¶ If True, sets strain to zero on open boundaries.
-
logical
freeslip_strain= .false.¶ If True, sets normal gradient of tangential velocity to zero in the strain on open boundaries.
-
logical
computed_strain= .false.¶ If True, uses external data for tangential velocity to compute normal gradient in the strain on open boundaries.
-
logical
specified_strain= .false.¶ If True, uses external data for tangential velocity gradients to compute strain on open boundaries.
-
logical
zero_biharmonic= .false.¶ If True, zeros the Laplacian of flow on open boundaries for use in the biharmonic viscosity term.
-
logical
brushcutter_mode= .false.¶ If True, read data on supergrid.
-
real
g_earth¶ The gravitational acceleration [m s-2].
-
type(obc_segment_type), dimension(:), pointer mom_open_boundary::ocean_obc_type::segment => NULL() List of segment objects.
-
integer, dimension(:,:), pointer mom_open_boundary::ocean_obc_type::segnum_u => NULL() Segment number of u-points.
-
integer, dimension(:,:), pointer mom_open_boundary::ocean_obc_type::segnum_v => NULL() Segment number of v-points.
-
real
gamma_uv¶ The relative weighting for the baroclinic radiation velocities (or speed of characteristics) at the new time level (1) or the running mean (0) for velocities. Valid values range from 0 to 1, with a default of 0.3.
-
real
rx_max¶ The maximum magnitude of the baroclinic radiation velocity (or speed of characteristics) [m s-1]. The default value is 10 m s-1.
-
logical
obc_pe¶ Is there an open boundary on this tile?
-
type(remapping_cs), pointer mom_open_boundary::ocean_obc_type::remap_cs ALE remapping control structure for segments only.
-
type(obc_registry_type), pointer mom_open_boundary::ocean_obc_type::obc_reg => NULL() Registry type for boundaries.
-
real, dimension(:,:,:), pointer mom_open_boundary::ocean_obc_type::rx_normal => NULL() Array storage for restarts.
-
real, dimension(:,:,:), pointer mom_open_boundary::ocean_obc_type::ry_normal => NULL() Array storage for restarts.
-
real, dimension(:,:,:), pointer mom_open_boundary::ocean_obc_type::cff_normal => NULL() Array storage for restarts.
-
real
silly_h¶ A silly value of thickness outside of the domain that can be used to test the independence of the OBCs to this external data [H ~> m or kg m-2].
-
real
silly_u¶ A silly value of velocity outside of the domain that can be used to test the independence of the OBCs to this external data [m s-1].
-
integer
- type
This type is used for communication with other components via the FMS coupler. The element names and types can be changed only with great deliberation, hence the persistnce of things like the cutsy element name “avg_kount”.
Public Members
-
type(domain2d) ocean_model_mod::ocean_public_type::domain The domain for the surface fields.
-
logical
is_ocean_pe¶ .true. on processors that run the ocean model.
-
character(len=32) ocean_model_mod::ocean_public_type::instance_name = '' A name that can be used to identify this instance of an ocean model, for example in ensembles when writing messages.
-
integer, dimension(:), pointer ocean_model_mod::ocean_public_type::pelist => NULL() The list of ocean PEs.
-
logical, dimension(:,:), pointer ocean_model_mod::ocean_public_type::maskmap =>NULL() A pointer to an array indicating which logical processors are actually used for the ocean code. The other logical processors would be all land points and are not assigned to actual processors. This need not be assigned if all logical processors are used.
-
integer
stagger= -999¶ The staggering relative to the tracer points points of the two velocity components. Valid entries include AGRID, BGRID_NE, CGRID_NE, BGRID_SW, and CGRID_SW, corresponding to the community-standard Arakawa notation. (These are named integers taken from mpp_parameter_mod.) Following MOM5, stagger is BGRID_NE by default when the ocean is initialized, but here it is set to -999 so that a global max across ocean and non-ocean processors can be used to determine its value.
-
real, dimension(:,:), pointer ocean_model_mod::ocean_public_type::t_surf => NULL() SST on t-cell (degrees Kelvin)
-
real, dimension(:,:), pointer ocean_model_mod::ocean_public_type::s_surf => NULL() SSS on t-cell (psu)
-
real, dimension(:,:), pointer ocean_model_mod::ocean_public_type::u_surf => NULL() i-velocity at the locations indicated by stagger [m s-1].
-
real, dimension(:,:), pointer ocean_model_mod::ocean_public_type::v_surf => NULL() j-velocity at the locations indicated by stagger [m s-1].
-
real, dimension(:,:), pointer ocean_model_mod::ocean_public_type::sea_lev => NULL() Sea level in m after correction for surface pressure,.
-
real, dimension(:,:), pointer ocean_model_mod::ocean_public_type::frazil =>NULL() Accumulated heating [J m-2] from frazil.
-
real, dimension(:,:), pointer ocean_model_mod::ocean_public_type::area => NULL() cell area of the ocean surface [m2].
-
type(coupler_2d_bc_type) ocean_model_mod::ocean_public_type::fields A structure that may contain named arrays of tracer-related surface fields.
-
integer
avg_kount¶ A count of contributions to running sums, used externally by the FMS coupler for accumulating averages of this type.
-
integer, dimension(2) ocean_model_mod::ocean_public_type::axes = 0 Axis numbers that are available for I/O using this surface data.
-
- type
The ocean_state_type contains all information about the state of the ocean, with a format that is private so it can be readily changed without disrupting other coupled components.
Public Members
-
logical
is_ocean_pe= .false.¶ True if this is an ocean PE.
-
type(time_type) ocean_model_mod::ocean_state_type::time The ocean model’s time and master clock.
-
type(time_type) ocean_model_mod::ocean_state_type::time_dyn The ocean model’s time for the dynamics. Time and Time_dyn should be the same after a full time step.
-
integer
restart_control¶ An integer that is bit-tested to determine whether incremental restart files are saved and whether they have a time stamped name. +1 (bit 0) for generic files and +2 (bit 1) for time-stamped files. A restart file is saved at the end of a run segment unless Restart_control is negative.
-
integer
nstep= 0¶ The number of calls to update_ocean that update the dynamics.
-
integer
nstep_thermo= 0¶ The number of calls to update_ocean that update the thermodynamics.
-
logical
use_ice_shelf¶ If true, the ice shelf model is enabled.
-
logical
use_waves¶ If true use wave coupling.
-
logical
icebergs_alter_ocean¶ If true, the icebergs can change ocean the ocean dynamics and forcing fluxes.
-
real
press_to_z¶ A conversion factor between pressure and ocean depth in m, usually 1/(rho_0*g) [m Pa-1].
-
real
c_p¶ The heat capacity of seawater [J degC-1 kg-1].
-
logical
offline_tracer_mode= .false.¶ If false, use the model in prognostic mode with the barotropic and baroclinic dynamics, thermodynamics, etc. stepped forward integrated in time. If true, all of the above are bypassed with all fields necessary to integrate only the tracer advection and diffusion equation read in from files stored from a previous integration of the prognostic model.
-
logical
single_step_call¶ If true, advance the state of MOM with a single step including both dynamics and thermodynamics. If false, the two phases are advanced with separate calls. The default is true.
-
real
dt¶ (baroclinic) dynamics time step [s]
-
real
dt_therm¶ thermodynamics time step [s]
-
logical
thermo_spans_coupling¶ If true, thermodynamic and tracer time steps can span multiple coupled time steps.
-
logical
diabatic_first¶ If true, apply diabatic and thermodynamic processes before time stepping the dynamics.
-
type(directories) ocean_model_mod::ocean_state_type::dirs A structure containing several relevant directory paths.
-
type(mech_forcing) ocean_model_mod::ocean_state_type::forces A structure with the driving mechanical surface forces.
-
type(forcing) ocean_model_mod::ocean_state_type::fluxes A structure containing pointers to the thermodynamic ocean forcing fields.
-
type(forcing) ocean_model_mod::ocean_state_type::flux_tmp A secondary structure containing pointers to the ocean forcing fields for when multiple coupled timesteps are taken per thermodynamic step.
-
type(surface) ocean_model_mod::ocean_state_type::sfc_state A structure containing pointers to the ocean surface state fields.
-
type(ocean_grid_type), pointer ocean_model_mod::ocean_state_type::grid => NULL() A pointer to a grid structure containing metrics.
-
type(verticalgrid_type), pointer ocean_model_mod::ocean_state_type::gv => NULL() A pointer to a structure containing information.
-
type(unit_scale_type), pointer ocean_model_mod::ocean_state_type::us => NULL() A pointer to a structure containing dimensional.
-
type(mom_control_struct), pointer ocean_model_mod::ocean_state_type::mom_csp => NULL() A pointer to the MOM control structure.
-
type(ice_shelf_cs), pointer ocean_model_mod::ocean_state_type::ice_shelf_csp => NULL() A pointer to the control structure for the.
-
type(marine_ice_cs), pointer ocean_model_mod::ocean_state_type::marine_ice_csp => NULL() A pointer to the control structure for the.
-
type(wave_parameters_cs), pointer ocean_model_mod::ocean_state_type::waves A structure containing pointers to the surface wave fields.
-
type(surface_forcing_cs), pointer ocean_model_mod::ocean_state_type::forcing_csp => NULL() A pointer to the MOM forcing control structure.
-
type(mom_restart_cs), pointer ocean_model_mod::ocean_state_type::restart_csp => NULL() A pointer set to the restart control structure.
-
type(diag_ctrl), pointer ocean_model_mod::ocean_state_type::diag => NULL() A pointer to the diagnostic regulatory structure.
-
logical
- type
The control structure for the OCMPI2_CFC tracer package.
Unnamed Group
-
real
a1_11¶ Coefficients used in the CFC11 and CFC12 solubility calculation.
-
real
a1_12¶ Coefficients used in the CFC11 and CFC12 solubility calculation.
-
real
a2_11¶ Coefficients used in the CFC11 and CFC12 solubility calculation.
-
real
a2_12¶ Coefficients used in the CFC11 and CFC12 solubility calculation.
-
real
a3_11¶ Coefficients used in the CFC11 and CFC12 solubility calculation.
-
real
a3_12¶ Coefficients used in the CFC11 and CFC12 solubility calculation.
-
real
a4_11¶ Coefficients used in the CFC11 and CFC12 solubility calculation.
-
real
a4_12¶ Coefficients used in the CFC11 and CFC12 solubility calculation.
-
real
d1_11¶ Coefficients used in the CFC11 and CFC12 solubility calculation.
-
real
d1_12¶ Coefficients used in the CFC11 and CFC12 solubility calculation.
-
real
d2_11¶ Coefficients used in the CFC11 and CFC12 solubility calculation.
-
real
d2_12¶ Coefficients used in the CFC11 and CFC12 solubility calculation.
-
real
d3_11¶ Coefficients used in the CFC11 and CFC12 solubility calculation.
-
real
d3_12¶ Coefficients used in the CFC11 and CFC12 solubility calculation.
-
real
d4_11¶ Coefficients used in the CFC11 and CFC12 solubility calculation.
-
real
d4_12¶ Coefficients used in the CFC11 and CFC12 solubility calculation.
-
real
e1_11¶ Coefficients used in the CFC11 and CFC12 solubility calculation.
-
real
e1_12¶ Coefficients used in the CFC11 and CFC12 solubility calculation.
-
real
e2_11¶ Coefficients used in the CFC11 and CFC12 solubility calculation.
-
real
e2_12¶ Coefficients used in the CFC11 and CFC12 solubility calculation.
-
real
e3_11¶ Coefficients used in the CFC11 and CFC12 solubility calculation.
-
real
e3_12¶ Coefficients used in the CFC11 and CFC12 solubility calculation.
-
real
cfc11_ic_val= 0.0¶ The initial value assigned to CFC11 [mol m-3].
-
real
cfc12_ic_val= 0.0¶ The initial value assigned to CFC12 [mol m-3].
-
real
cfc11_land_val= -1.0¶ The value of CFC11 used where land is masked out [mol m-3].
-
real
cfc12_land_val= -1.0¶ The value of CFC12 used where land is masked out [mol m-3].
-
logical
tracers_may_reinit¶ If true, tracers may be reset via the initialization code if they are not found in the restart files.
-
character(len=16) mom_ocmip2_cfc::ocmip2_cfc_cs::cfc11_name CFC11 variable name.
-
character(len=16) mom_ocmip2_cfc::ocmip2_cfc_cs::cfc12_name CFC12 variable name.
-
integer
ind_cfc_11_flux¶ Index returned by aof_set_coupler_flux that is used to pack and unpack surface boundary condition arrays.
-
integer
ind_cfc_12_flux¶ Index returned by aof_set_coupler_flux that is used to pack and unpack surface boundary condition arrays.
-
type(diag_ctrl), pointer mom_ocmip2_cfc::ocmip2_cfc_cs::diag => NULL() Coefficients used in the CFC11 and CFC12 solubility calculation.
-
type(mom_restart_cs), pointer mom_ocmip2_cfc::ocmip2_cfc_cs::restart_csp => NULL() Coefficients used in the CFC11 and CFC12 solubility calculation.
-
type(vardesc) mom_ocmip2_cfc::ocmip2_cfc_cs::cfc11_desc A set of metadata for the CFC11 tracer.
-
type(vardesc) mom_ocmip2_cfc::ocmip2_cfc_cs::cfc12_desc A set of metadata for the CFC12 tracer.
Public Members
-
character(len=200) mom_ocmip2_cfc::ocmip2_cfc_cs::ic_file The file in which the CFC initial values can be found, or an empty string for internal initilaization.
-
logical
z_ic_file¶ If true, the IC_file is in Z-space. The default is false..
-
type(time_type), pointer mom_ocmip2_cfc::ocmip2_cfc_cs::time => NULL() A pointer to the ocean model’s clock.
-
type(tracer_registry_type), pointer mom_ocmip2_cfc::ocmip2_cfc_cs::tr_reg => NULL() A pointer to the MOM6 tracer registry.
-
real, dimension(:,:,:), pointer mom_ocmip2_cfc::ocmip2_cfc_cs::cfc11 => NULL() The CFC11 concentration [mol m-3].
-
real, dimension(:,:,:), pointer mom_ocmip2_cfc::ocmip2_cfc_cs::cfc12 => NULL() The CFC12 concentration [mol m-3].
-
real
- type
Control structure that contains a transpose of the ocean state across ensemble members.
Public Members
-
type(ocean_control_struct), pointer mom_oda_driver_mod::oda_cs::ocean_prior => NULL() ensemble ocean prior states in DA space
-
type(ocean_control_struct), pointer mom_oda_driver_mod::oda_cs::ocean_posterior => NULL() ensemble ocean posterior states or increments to prior in DA space
-
integer
nk¶ number of vertical layers used for DA
-
type(ocean_grid_type), pointer mom_oda_driver_mod::oda_cs::grid => NULL() MOM6 grid type and decomposition for the DA.
-
type(ptr_mpp_domain), dimension(:), pointer mom_oda_driver_mod::oda_cs::domains => NULL() Pointer to mpp_domain objects for ensemble members.
-
type(verticalgrid_type), pointer mom_oda_driver_mod::oda_cs::gv => NULL() vertical grid for DA
-
type(unit_scale_type), pointer mom_oda_driver_mod::oda_cs::us => NULL() structure containing various unit conversion factors for DA
-
type(domain2d), pointer mom_oda_driver_mod::oda_cs::mpp_domain => NULL() Pointer to a mpp domain object for DA.
-
type(grid_type), pointer mom_oda_driver_mod::oda_cs::oda_grid local tracer grid
-
real, dimension(:,:,:), pointer mom_oda_driver_mod::oda_cs::h => NULL() layer thicknesses [H ~> m or kg m-2] for DA
-
type(thermo_var_ptrs), pointer mom_oda_driver_mod::oda_cs::tv => NULL() pointer to thermodynamic variables
-
integer
ni¶ global i-direction grid size
-
integer
nj¶ global j-direction grid size
-
logical
reentrant_x¶ grid is reentrant in the x direction
-
logical
reentrant_y¶ grid is reentrant in the y direction
-
logical
tripolar_n¶ grid is folded at its north edge
-
logical
symmetric¶ Values at C-grid locations are symmetric.
-
integer
assim_method¶ Method: NO_ASSIM,EAKF_ASSIM or OI_ASSIM.
-
integer
ensemble_size¶ Size of the ensemble.
-
integer
ensemble_id= 0¶ id of the current ensemble member
-
integer, dimension(:,:), pointer mom_oda_driver_mod::oda_cs::ensemble_pelist PE list for ensemble members.
-
integer, dimension(:), pointer mom_oda_driver_mod::oda_cs::filter_pelist PE list for ensemble members.
-
integer
assim_frequency¶ analysis interval in hours
-
type(ocean_profile_type), pointer mom_oda_driver_mod::oda_cs::profiles => NULL() pointer to linked list of all available profiles
-
type(ocean_profile_type), pointer mom_oda_driver_mod::oda_cs::cprofiles => NULL() pointer to linked list of current profiles
-
type(kd_root), pointer mom_oda_driver_mod::oda_cs::kdroot => NULL() A structure for storing nearest neighbors.
-
type(ale_cs), pointer mom_oda_driver_mod::oda_cs::ale_cs =>NULL() ALE control structure for DA.
-
logical
use_ale_algorithm¶ true is using ALE remapping
-
type(regridding_cs) mom_oda_driver_mod::oda_cs::regridcs ALE control structure for regridding.
-
type(remapping_cs) mom_oda_driver_mod::oda_cs::remapcs ALE control structure for remapping.
-
type(time_type) mom_oda_driver_mod::oda_cs::time Current Analysis time.
-
type(diag_ctrl) mom_oda_driver_mod::oda_cs::diag_cs Diagnostics control structure.
-
- type
The control structure for the offline transport module.
Unnamed Group
-
integer
id_uhr= -1¶ Diagnostic manager IDs for some fields that may be of interest when doing offline transport.
-
integer
id_vhr= -1¶ Diagnostic manager IDs for some fields that may be of interest when doing offline transport.
-
integer
id_ear= -1¶ Diagnostic manager IDs for some fields that may be of interest when doing offline transport.
-
integer
id_ebr= -1¶ Diagnostic manager IDs for some fields that may be of interest when doing offline transport.
-
integer
id_hr= -1¶ Diagnostic manager IDs for some fields that may be of interest when doing offline transport.
-
integer
id_hdiff= -1¶ Diagnostic manager IDs for some fields that may be of interest when doing offline transport.
-
integer
id_uhr_redist= -1¶ Diagnostic manager IDs for some fields that may be of interest when doing offline transport.
-
integer
id_vhr_redist= -1¶ Diagnostic manager IDs for some fields that may be of interest when doing offline transport.
-
integer
id_uhr_end= -1¶ Diagnostic manager IDs for some fields that may be of interest when doing offline transport.
-
integer
id_vhr_end= -1¶ Diagnostic manager IDs for some fields that may be of interest when doing offline transport.
-
integer
id_eta_pre_distribute= -1¶ Diagnostic manager IDs for some fields that may be of interest when doing offline transport.
-
integer
id_eta_post_distribute= -1¶ Diagnostic manager IDs for some fields that may be of interest when doing offline transport.
-
integer
id_h_redist= -1¶ Diagnostic manager IDs for some fields that may be of interest when doing offline transport.
-
integer
id_eta_diff_end= -1¶ Diagnostic manager IDs for some fields that may be of interest when doing offline transport.
-
integer
id_uhtr_regrid= -1¶ Diagnostic manager IDs for some fields that may be of interest when doing offline transport.
-
integer
id_vhtr_regrid= -1¶ Diagnostic manager IDs for some fields that may be of interest when doing offline transport.
-
integer
id_temp_regrid= -1¶ Diagnostic manager IDs for some fields that may be of interest when doing offline transport.
-
integer
id_salt_regrid= -1¶ Diagnostic manager IDs for some fields that may be of interest when doing offline transport.
-
integer
id_h_regrid= -1¶ Diagnostic manager IDs for some fields that may be of interest when doing offline transport.
-
integer
id_clock_read_fields= -1¶ A CPU time clock.
-
integer
id_clock_offline_diabatic= -1¶ A CPU time clock.
-
integer
id_clock_offline_adv= -1¶ A CPU time clock.
-
integer
id_clock_redistribute= -1¶ A CPU time clock.
-
real, dimension(:,:,:), allocatable mom_offline_main::offline_transport_cs::uhtr Zonal transport that may need to be stored between calls to step_MOM.
-
real, dimension(:,:,:), allocatable mom_offline_main::offline_transport_cs::vhtr Meridional transport that may need to be stored between calls to step_MOM.
-
real, dimension(:,:,:), allocatable mom_offline_main::offline_transport_cs::eatr Amount of fluid entrained from the layer above within one time step [H ~> m or kg m-2].
-
real, dimension(:,:,:), allocatable mom_offline_main::offline_transport_cs::ebtr Amount of fluid entrained from the layer below within one time step [H ~> m or kg m-2].
-
real, dimension(:,:,:), allocatable mom_offline_main::offline_transport_cs::kd Vertical diffusivity.
-
real, dimension(:,:,:), allocatable mom_offline_main::offline_transport_cs::h_end Thicknesses at the end of offline timestep.
-
real, dimension(:,:), allocatable mom_offline_main::offline_transport_cs::netmassin Freshwater fluxes into the ocean.
-
real, dimension(:,:), allocatable mom_offline_main::offline_transport_cs::netmassout Freshwater fluxes out of the ocean.
-
real, dimension(:,:), allocatable mom_offline_main::offline_transport_cs::mld Mixed layer depths at thickness points [H ~> m or kg m-2].
-
real, dimension(:,:,:,:), allocatable mom_offline_main::offline_transport_cs::uhtr_all Entire field of zonal transport.
-
real, dimension(:,:,:,:), allocatable mom_offline_main::offline_transport_cs::vhtr_all Entire field of mericional transport.
-
real, dimension(:,:,:,:), allocatable mom_offline_main::offline_transport_cs::hend_all Entire field of layer thicknesses.
-
real, dimension(:,:,:,:), allocatable mom_offline_main::offline_transport_cs::temp_all Entire field of temperatures.
-
real, dimension(:,:,:,:), allocatable mom_offline_main::offline_transport_cs::salt_all Entire field of salinities.
Public Members
-
type(ale_cs), pointer mom_offline_main::offline_transport_cs::ale_csp => NULL() A pointer to the ALE control structure.
-
type(diabatic_cs), pointer mom_offline_main::offline_transport_cs::diabatic_csp => NULL() A pointer to the diabatic control structure.
-
type(diag_ctrl), pointer mom_offline_main::offline_transport_cs::diag => NULL() Structure that regulates diagnostic output.
-
type(ocean_obc_type), pointer mom_offline_main::offline_transport_cs::obc => NULL() A pointer to the open boundary condition control structure.
-
type(tracer_advect_cs), pointer mom_offline_main::offline_transport_cs::tracer_adv_csp => NULL() A pointer to the tracer advection control structure.
-
type(opacity_cs), pointer mom_offline_main::offline_transport_cs::opacity_csp => NULL() A pointer to the opacity control structure.
-
type(tracer_flow_control_cs), pointer mom_offline_main::offline_transport_cs::tracer_flow_csp => NULL() A pointer to control structure that orchestrates the calling of tracer packages.
-
type(tracer_registry_type), pointer mom_offline_main::offline_transport_cs::tracer_reg => NULL() A pointer to the tracer registry.
-
type(thermo_var_ptrs), pointer mom_offline_main::offline_transport_cs::tv => NULL() A structure pointing to various thermodynamic variables.
-
type(ocean_grid_type), pointer mom_offline_main::offline_transport_cs::g => NULL() Pointer to a structure containing metrics and related information.
-
type(verticalgrid_type), pointer mom_offline_main::offline_transport_cs::gv => NULL() Pointer to structure containing information about the vertical grid.
-
type(optics_type), pointer mom_offline_main::offline_transport_cs::optics => NULL() Pointer to the optical properties type.
-
type(diabatic_aux_cs), pointer mom_offline_main::offline_transport_cs::diabatic_aux_csp => NULL() Pointer to the diabatic_aux control structure.
-
integer
start_index¶ Variables related to reading in fields from online run.
Timelevel to start
-
integer
iter_no¶ Timelevel to start.
-
integer
numtime¶ How many timelevels in the input fields.
-
integer
accumulated_time¶ Length of time accumulated in the current offline interval.
-
integer
ridx_sum= -1¶ Read index offset of the summed variables.
-
integer
ridx_snap= -1¶ Read index offset of the snapshot variables.
-
integer
nk_input¶ Number of input levels in the input fields.
-
character(len=200) mom_offline_main::offline_transport_cs::offlinedir Directory where offline fields are stored.
-
character(len=200) mom_offline_main::offline_transport_cs::surf_file Contains surface fields (2d arrays)
-
character(len=200) mom_offline_main::offline_transport_cs::snap_file Snapshotted fields (layer thicknesses)
-
character(len=200) mom_offline_main::offline_transport_cs::sum_file Fields which are accumulated over time.
-
character(len=200) mom_offline_main::offline_transport_cs::mean_file Fields averaged over time.
-
character(len=20) mom_offline_main::offline_transport_cs::redistribute_method ‘barotropic’ if evenly distributing extra flow throughout entire watercolumn, ‘upwards’, if trying to do it just in the layers above ‘both’ if both methods are used
-
character(len=20) mom_offline_main::offline_transport_cs::mld_var_name Name of the mixed layer depth variable to use.
-
logical
fields_are_offset¶ True if the time-averaged fields and snapshot fields are offset by one time level.
-
logical
x_before_y¶ Which horizontal direction is advected first.
-
logical
print_adv_offline¶ Prints out some updates each advection sub interation.
-
logical
skip_diffusion¶ Skips horizontal diffusion of tracers.
-
logical
read_sw¶ Read in averaged values for shortwave radiation.
-
logical
read_mld¶ Check to see whether mixed layer depths should be read in.
-
logical
diurnal_sw¶ Adds a synthetic diurnal cycle on shortwave radiation.
-
logical
debug¶ If true, write verbose debugging messages.
-
logical
redistribute_barotropic¶ Redistributes column-summed residual transports throughout a column weighted by thickness.
-
logical
redistribute_upwards¶ Redistributes remaining fluxes only in layers above the current one based as the max allowable transport in that cell.
-
logical
read_all_ts_uvh¶ If true, then all timelevels of temperature, salinity, mass transports, and Layer thicknesses are read during initialization Variables controlling some of the numerical considerations of offline transport.
-
integer
num_off_iter¶ Number of advection iterations per offline step.
-
integer
num_vert_iter¶ Number of vertical iterations per offline step.
-
integer
off_ale_mod¶ Sets how frequently the ALE step is done during the advection.
-
real
dt_offline¶ Timestep used for offline tracers [s].
-
real
dt_offline_vertical¶ Timestep used for calls to tracer vertical physics [s].
-
real
evap_cfl_limit¶ Copied from diabatic_CS controlling how tracers follow freshwater fluxes.
-
real
minimum_forcing_depth¶ Copied from diabatic_CS controlling how tracers follow freshwater fluxes.
-
real
kd_max¶ Runtime parameter specifying the maximum value of vertical diffusivity.
-
real
min_residual¶ The minimum amount of total mass flux before exiting the main advection routine.
-
integer
- type
The control structure for the oil tracer package.
Public Members
-
integer
ntr¶ The number of tracers that are actually used.
-
logical
coupled_tracers= .false.¶ These tracers are not offered to the coupler.
-
character(len=200) oil_tracer::oil_tracer_cs::ic_file The file in which the age-tracer initial values can be found, or an empty string for internal initialization.
-
logical
z_ic_file¶ If true, the IC_file is in Z-space. The default is false.
-
real
oil_source_longitude¶ Latitude of source location (geographic)
-
real
oil_source_latitude¶ Longitude of source location (geographic)
-
integer
oil_source_i= -999¶ Local i of source location (computational)
-
integer
oil_source_j= -999¶ Local j of source location (computational)
-
real
oil_source_rate¶ Rate of oil injection [kg s-1].
-
real
oil_start_year¶ The year in which tracers start aging, or at which the surface value equals young_val, in years.
-
real
oil_end_year¶ The year in which tracers start aging, or at which the surface value equals young_val, in years.
-
type(time_type), pointer oil_tracer::oil_tracer_cs::time => NULL() A pointer to the ocean model’s clock.
-
type(tracer_registry_type), pointer oil_tracer::oil_tracer_cs::tr_reg => NULL() A pointer to the MOM tracer registry.
-
real, dimension(:,:,:,:), pointer oil_tracer::oil_tracer_cs::tr => NULL() The array of tracers used in this subroutine, in g m-3?
-
real, dimension(ntr_max) oil_tracer::oil_tracer_cs::ic_val = 0.0 The (uniform) initial condition value.
-
real, dimension(ntr_max) oil_tracer::oil_tracer_cs::young_val = 0.0 The value assigned to tr at the surface.
-
real, dimension(ntr_max) oil_tracer::oil_tracer_cs::land_val = -1.0 The value of tr used where land is masked out.
-
real, dimension(ntr_max) oil_tracer::oil_tracer_cs::sfc_growth_rate The exponential growth rate for the surface value [year-1].
-
real, dimension(ntr_max) oil_tracer::oil_tracer_cs::oil_decay_days Decay time scale of oil [days].
-
real, dimension(ntr_max) oil_tracer::oil_tracer_cs::oil_decay_rate Decay rate of oil [s-1] calculated from oil_decay_days.
-
integer, dimension(ntr_max) oil_tracer::oil_tracer_cs::oil_source_k Layer of source.
-
logical
oil_may_reinit¶ If true, oil tracers may be reset by the initialization code if they are not found in the restart files.
-
integer, dimension(ntr_max) oil_tracer::oil_tracer_cs::ind_tr Indices returned by aof_set_coupler_flux if it is used and the surface tracer concentrations are to be provided to the coupler.
-
type(vardesc), dimension(ntr_max) oil_tracer::oil_tracer_cs::tr_desc Descriptions and metadata for the tracers.
-
type(diag_ctrl), pointer oil_tracer::oil_tracer_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
type(mom_restart_cs), pointer oil_tracer::oil_tracer_cs::restart_csp => NULL() A pointer to the restart control structure.
-
integer
- type
The control structure with paramters for the MOM_opacity module.
Unnamed Group
-
integer
id_sw_pen= -1¶ Diagnostic IDs.
-
integer
id_sw_vis_pen= -1¶ Diagnostic IDs.
-
integer, dimension(:), pointer mom_opacity::opacity_cs::id_opacity => NULL() Diagnostic IDs.
Public Members
-
logical
var_pen_sw¶ If true, use one of the CHL_A schemes (specified by OPACITY_SCHEME) to determine the e-folding depth of incoming shortwave radiation.
-
integer
opacity_scheme¶ An integer indicating which scheme should be used to translate water properties into the opacity (i.e., the e-folding depth) and (perhaps) the number of bands of penetrating shortwave radiation to use.
-
real
pen_sw_scale¶ The vertical absorption e-folding depth of the penetrating shortwave radiation [m].
-
real
pen_sw_scale_2nd¶ The vertical absorption e-folding depth of the (2nd) penetrating shortwave radiation [m].
-
real
sw_1st_exp_ratio¶ Ratio for 1st exp decay in Two Exp decay opacity.
-
real
pen_sw_frac¶ The fraction of shortwave radiation that is penetrating with a constant e-folding approach.
-
real
blue_frac¶ The fraction of the penetrating shortwave radiation that is in the blue band [nondim].
-
real
opacity_land_value¶ The value to use for opacity over land [m-1]. The default is 10 m-1 - a value for muddy water.
-
type(diag_ctrl), pointer mom_opacity::opacity_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
integer
- interface
Add two extended-fixed-point numbers.
Private Functions
-
type(efp_type) function mom_coms::operator(+)::efp_plus(EFP1 EFP1, EFP2 EFP2) Add two extended-fixed-point numbers.
- Return
The result in extended fixed point format
- Parameters
[in] efp1: The first extended fixed point number[in] efp2: The second extended fixed point number
-
- interface
Subtract one extended-fixed-point number from another.
Private Functions
-
type(efp_type) function mom_coms::operator(-)::efp_minus(EFP1 EFP1, EFP2 EFP2) Subract one extended-fixed-point number from another.
- Return
The result in extended fixed point format
- Parameters
[in] efp1: The first extended fixed point number[in] efp2: The extended fixed point number being subtracted from the first extended fixed point number
-
- type
This type is used to store information about ocean optical properties.
Public Members
-
integer
nbands¶ The number of penetrating bands of SW radiation.
-
real, dimension(:,:,:,:), pointer mom_opacity::optics_type::opacity_band => NULL() SW optical depth per unit thickness [m-1] The number of radiation bands is most rapidly varying (first) index.
-
real, dimension(:,:,:), pointer mom_opacity::optics_type::sw_pen_band => NULL() shortwave radiation [W m-2] at the surface in each of the nbands bands that penetrates beyond the surface. The most rapidly varying dimension is the band.
-
real, dimension(:), pointer mom_opacity::optics_type::min_wavelength_band => NULL() The minimum wavelength in each band of penetrating shortwave radiation [nm].
-
real, dimension(:), pointer mom_opacity::optics_type::max_wavelength_band => NULL() The maximum wavelength in each band of penetrating shortwave radiation [nm].
-
real
pensw_flux_absorb¶ A heat flux that is small enough to be completely absorbed in the next sufficiently thick layer [H degC T-1 ~> degC m s-1 or degC kg m-2 s-1].
-
real
pensw_absorb_invlen¶ The inverse of the thickness that is used to absorb the remaining shortwave heat flux when it drops below PEN_SW_FLUX_ABSORB [H ~> m or kg m-2].
-
logical
answers_2018¶ If true, use the order of arithmetic and expressions that recover the answers from the end of 2018. Otherwise, use updated and more robust forms of the same expressions.
-
integer
- type
A type for making arrays of pointers to scalars.
Private Members
-
real, pointer mom_restart::p0d::p => NULL() A pointer to a scalar.
-
- type
A type for making arrays of pointers to 1-d arrays.
Private Members
-
real, dimension(:), pointer mom_restart::p1d::p => NULL() A pointer to a 1d array.
-
- type
A type for making arrays of pointers to 2-d arrays.
Private Members
-
real, dimension(:,:), pointer mom_restart::p2d::p => NULL() A pointer to a 2d array.
-
- type
A type that can be used to create arrays of pointers to 2D arrays.
Unnamed Group
-
real, dimension(:,:), pointer mom_tracer_hor_diff::p2d::p => NULL() A pointer to a 2D array of reals.
-
- type
A structure for creating arrays of pointers to 2D arrays with extra gridding information.
Private Members
-
integer
id¶ id for FMS external time interpolator
-
integer
nz_data¶ The number of vertical levels in the input field.
-
integer
num_tlevs¶ The number of time records contained in the file.
-
real, dimension(:,:), pointer mom_ale_sponge::p2d::mask_in => NULL() pointer to the data mask.
-
real, dimension(:,:), pointer mom_ale_sponge::p2d::p => NULL() pointer the data.
-
real, dimension(:,:), pointer mom_ale_sponge::p2d::h => NULL() pointer the data grid.
-
integer
- type
A structure for creating arrays of pointers to 2D arrays.
Public Members
-
real, dimension(:,:), pointer mom_sponge::p2d::p => NULL() A pointer to a 2D array.
-
- type
A structure for creating arrays of pointers to 2D arrays.
Public Members
-
real, dimension(:,:), pointer mom_variables::p2d::p => NULL() A pointer to a 2D array.
-
- type
A type that can be used to create arrays of pointers to 2D integer arrays.
Unnamed Group
-
integer, dimension(:,:), pointer mom_tracer_hor_diff::p2di::p => NULL() A pointer to a 2D array of integers.
-
- type
A structure for creating arrays of pointers to 3D arrays.
Public Members
-
real, dimension(:,:,:), pointer mom_sponge::p3d::p => NULL() A pointer to a 3D array.
-
- type
A type for making arrays of pointers to 3-d arrays.
Private Members
-
real, dimension(:,:,:), pointer mom_restart::p3d::p => NULL() A pointer to a 3d array.
-
- type
A structure for creating arrays of pointers to 3D arrays.
Public Members
-
real, dimension(:,:,:), pointer mom_variables::p3d::p => NULL() A pointer to a 3D array.
-
- type
A structure for creating arrays of pointers to 3D arrays with extra gridding information.
Private Members
-
integer
id¶ id for FMS external time interpolator
-
integer
nz_data¶ The number of vertical levels in the input field.
-
integer
num_tlevs¶ The number of time records contained in the file.
-
real, dimension(:,:,:), pointer mom_ale_sponge::p3d::mask_in => NULL() pointer to the data mask.
-
real, dimension(:,:,:), pointer mom_ale_sponge::p3d::p => NULL() pointer to the data.
-
real, dimension(:,:,:), pointer mom_ale_sponge::p3d::h => NULL() pointer to the data grid.
-
integer
- type
A type for making arrays of pointers to 4-d arrays.
Private Members
-
real, dimension(:,:,:,:), pointer mom_restart::p4d::p => NULL() A pointer to a 4d array.
-
- type
A structure that can be parsed to read and document run-time parameters.
Unnamed Group
-
integer
nfiles= 0¶ The number of open files.
-
integer, dimension(max_param_files) mom_file_parser::param_file_type::iounit The unit numbers of open files.
-
character(len=filename_length), dimension(max_param_files) mom_file_parser::param_file_type::filename The names of the open files.
-
logical, dimension(max_param_files) mom_file_parser::param_file_type::netcdf_file If true, the input file is in NetCDF.
-
type(file_data_type), dimension(max_param_files) mom_file_parser::param_file_type::param_data Structures that contain the valid data lines from the parameter files, enabling all subsequent reads of parameter data to occur internally.
-
logical
report_unused= report_unused_default¶ If true, report any parameter lines that are not used in the run.
-
logical
unused_params_fatal= unused_params_fatal_default¶ If true, kill the run if there are any unused parameters.
-
logical
log_to_stdout= log_to_stdout_default¶ If true, all log messages are also sent to stdout.
-
logical
log_open= .false.¶ True if the log file has been opened.
-
character(len=240) mom_file_parser::param_file_type::doc_file A file where all run-time parameters, their settings and defaults are documented.
-
logical
complete_doc= complete_doc_default¶ If true, document all run-time parameters.
-
logical
minimal_doc= minimal_doc_default¶ If true, document only those run-time parameters that differ from defaults.
-
type(doc_type), pointer mom_file_parser::param_file_type::doc => NULL() A structure that contains information related to parameter documentation.
-
type(link_parameter), pointer mom_file_parser::param_file_type::chain => NULL() Facilitates linked list.
-
type(parameter_block), pointer mom_file_parser::param_file_type::blockname => NULL() Name of active parameter block.
-
integer
- type
Specify the active parameter block.
Unnamed Group
-
character(len=240) mom_file_parser::parameter_block::name = '' The active parameter block name.
-
-
interface
pass_var¶ Do a halo update on an array.
Private Functions
-
subroutine
pass_var_3d(array array, MOM_dom MOM_dom, sideflag sideflag, complete complete, position position, halo halo, clock clock)¶ pass_var_3d does a halo update for a three-dimensional array.
- Parameters
[inout] array: The array which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] sideflag: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH. For example, TO_EAST sends the data to the processor to the east, sothe halos on the western side are filled. TO_ALL is the default if sideflag is omitted.[in] complete: An optional argument indicating whether the halo updates should be completed before progress resumes. Omitting complete is the same as setting complete to .true.[in] position: An optional argument indicating the position. This is usally CORNER, but is CENTER by default.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
pass_var_2d(array array, MOM_dom MOM_dom, sideflag sideflag, complete complete, position position, halo halo, inner_halo inner_halo, clock clock)¶ pass_var_2d does a halo update for a two-dimensional array.
- Parameters
[inout] array: The array which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] sideflag: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if sideflag is omitted.[in] complete: An optional argument indicating whether the halo updates should be completed before progress resumes. Omitting complete is the same as setting complete to .true.[in] position: An optional argument indicating the position. This is usally CORNER, but is CENTER by default.[in] halo: The size of the halo to update - the full halo by default.[in] inner_halo: The size of an inner halo to avoid updating, or 0 to avoid updating symmetric memory computational domain points. Setting this >=0 also enforces that complete=.true.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
-
interface
pass_var_complete¶ Complete a non-blocking halo update on an array.
Private Functions
-
subroutine
pass_var_complete_3d(id_update id_update, array array, MOM_dom MOM_dom, sideflag sideflag, position position, halo halo, clock clock)¶ pass_var_complete_3d completes a halo update for a three-dimensional array.
- Parameters
[in] id_update: The integer id of this update which has been returned from a previous call to pass_var_start.[inout] array: The array which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] sideflag: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if sideflag is omitted.[in] position: An optional argument indicating the position. This is usally CORNER, but is CENTER by default.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
pass_var_complete_2d(id_update id_update, array array, MOM_dom MOM_dom, sideflag sideflag, position position, halo halo, clock clock)¶ pass_var_complete_2d completes a halo update for a two-dimensional array.
- Parameters
[in] id_update: The integer id of this update which has been returned from a previous call to pass_var_start.[inout] array: The array which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] sideflag: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if sideflag is omitted.[in] position: An optional argument indicating the position. This is usally CORNER, but is CENTER by default.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
-
interface
pass_var_start¶ Initiate a non-blocking halo update on an array.
Private Functions
-
integer function mom_domains::pass_var_start::pass_var_start_3d(array array, MOM_dom MOM_dom, sideflag sideflag, position position, complete complete, halo halo, clock clock) pass_var_start_3d starts a halo update for a three-dimensional array.
- Return
The integer index for this update.
- Parameters
[inout] array: The array which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] sideflag: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if sideflag is omitted.[in] position: An optional argument indicating the position. This is usally CORNER, but is CENTER by default.[in] complete: An optional argument indicating whether the halo updates should be completed before progress resumes. Omitting complete is the same as setting complete to .true.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
integer function mom_domains::pass_var_start::pass_var_start_2d(array array, MOM_dom MOM_dom, sideflag sideflag, position position, complete complete, halo halo, clock clock) pass_var_start_2d starts a halo update for a two-dimensional array.
- Return
The integer index for this update.
- Parameters
[inout] array: The array which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] sideflag: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if sideflag is omitted.[in] position: An optional argument indicating the position. This is usally CORNER, but is CENTER by default.[in] complete: An optional argument indicating whether the halo updates should be completed before progress resumes. Omitting complete is the same as setting complete to .true.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
-
interface
pass_vector¶ Do a halo update on a pair of arrays representing the two components of a vector.
Private Functions
-
subroutine
pass_vector_3d(u_cmpt u_cmpt, v_cmpt v_cmpt, MOM_dom MOM_dom, direction direction, stagger stagger, complete complete, halo halo, clock clock)¶ pass_vector_3d does a halo update for a pair of three-dimensional arrays representing the compontents of a three-dimensional horizontal vector.
- Parameters
[inout] u_cmpt: The nominal zonal (u) component of the vector pair which is having its halos points exchanged.[inout] v_cmpt: The nominal meridional (v) component of the vector pair which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] direction: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH, possibly plus SCALAR_PAIR if these are paired non-directional scalars discretized at the typical vector component locations. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if omitted.[in] stagger: An optional flag, which may be one of A_GRID, BGRID_NE, or CGRID_NE, indicating where the two components of the vector are discretized. Omitting stagger is the same as setting it to CGRID_NE.[in] complete: An optional argument indicating whether the halo updates should be completed before progress resumes. Omitting complete is the same as setting complete to .true.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
pass_vector_2d(u_cmpt u_cmpt, v_cmpt v_cmpt, MOM_dom MOM_dom, direction direction, stagger stagger, complete complete, halo halo, clock clock)¶ pass_vector_2d does a halo update for a pair of two-dimensional arrays representing the compontents of a two-dimensional horizontal vector.
- Parameters
[inout] u_cmpt: The nominal zonal (u) component of the vector pair which is having its halos points exchanged.[inout] v_cmpt: The nominal meridional (v) component of the vector pair which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] direction: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH, possibly plus SCALAR_PAIR if these are paired non-directional scalars discretized at the typical vector component locations. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if omitted.[in] stagger: An optional flag, which may be one of A_GRID, BGRID_NE, or CGRID_NE, indicating where the two components of the vector are discretized. Omitting stagger is the same as setting it to CGRID_NE.[in] complete: An optional argument indicating whether the halo updates should be completed before progress resumes. Omitting complete is the same as setting complete to .true.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
-
interface
pass_vector_complete¶ Complete a halo update on a pair of arrays representing the two components of a vector.
Private Functions
-
subroutine
pass_vector_complete_3d(id_update id_update, u_cmpt u_cmpt, v_cmpt v_cmpt, MOM_dom MOM_dom, direction direction, stagger stagger, halo halo, clock clock)¶ pass_vector_complete_3d completes a halo update for a pair of three-dimensional arrays representing the compontents of a three-dimensional horizontal vector.
- Parameters
[in] id_update: The integer id of this update which has been returned from a previous call to pass_var_start.[inout] u_cmpt: The nominal zonal (u) component of the vector pair which is having its halos points exchanged.[inout] v_cmpt: The nominal meridional (v) component of the vector pair which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] direction: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH, possibly plus SCALAR_PAIR if these are paired non-directional scalars discretized at the typical vector component locations. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if omitted.[in] stagger: An optional flag, which may be one of A_GRID, BGRID_NE, or CGRID_NE, indicating where the two components of the vector are discretized. Omitting stagger is the same as setting it to CGRID_NE.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
pass_vector_complete_2d(id_update id_update, u_cmpt u_cmpt, v_cmpt v_cmpt, MOM_dom MOM_dom, direction direction, stagger stagger, halo halo, clock clock)¶ pass_vector_complete_2d completes a halo update for a pair of two-dimensional arrays representing the compontents of a two-dimensional horizontal vector.
- Parameters
[in] id_update: The integer id of this update which has been returned from a previous call to pass_var_start.[inout] u_cmpt: The nominal zonal (u) component of the vector pair which is having its halos points exchanged.[inout] v_cmpt: The nominal meridional (v) component of the vector pair which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] direction: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH, possibly plus SCALAR_PAIR if these are paired non-directional scalars discretized at the typical vector component locations. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if omitted.[in] stagger: An optional flag, which may be one of A_GRID, BGRID_NE, or CGRID_NE, indicating where the two components of the vector are discretized. Omitting stagger is the same as setting it to CGRID_NE.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
-
interface
pass_vector_start¶ Initiate a halo update on a pair of arrays representing the two components of a vector.
Private Functions
-
integer function mom_domains::pass_vector_start::pass_vector_start_3d(u_cmpt u_cmpt, v_cmpt v_cmpt, MOM_dom MOM_dom, direction direction, stagger stagger, complete complete, halo halo, clock clock) pass_vector_start_3d starts a halo update for a pair of three-dimensional arrays representing the compontents of a three-dimensional horizontal vector.
- Return
The integer index for this update.
- Parameters
[inout] u_cmpt: The nominal zonal (u) component of the vector pair which is having its halos points exchanged.[inout] v_cmpt: The nominal meridional (v) component of the vector pair which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] direction: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH, possibly plus SCALAR_PAIR if these are paired non-directional scalars discretized at the typical vector component locations. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if omitted.[in] stagger: An optional flag, which may be one of A_GRID, BGRID_NE, or CGRID_NE, indicating where the two components of the vector are discretized. Omitting stagger is the same as setting it to CGRID_NE.[in] complete: An optional argument indicating whether the halo updates should be completed before progress resumes. Omitting complete is the same as setting complete to .true.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
integer function mom_domains::pass_vector_start::pass_vector_start_2d(u_cmpt u_cmpt, v_cmpt v_cmpt, MOM_dom MOM_dom, direction direction, stagger stagger, complete complete, halo halo, clock clock) pass_vector_start_2d starts a halo update for a pair of two-dimensional arrays representing the compontents of a two-dimensional horizontal vector.
- Return
The integer index for this update.
- Parameters
[inout] u_cmpt: The nominal zonal (u) component of the vector pair which is having its halos points exchanged.[inout] v_cmpt: The nominal meridional (v) component of the vector pair which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] direction: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH, possibly plus SCALAR_PAIR if these are paired non-directional scalars discretized at the typical vector component locations. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if omitted.[in] stagger: An optional flag, which may be one of A_GRID, BGRID_NE, or CGRID_NE, indicating where the two components of the vector are discretized. Omitting stagger is the same as setting it to CGRID_NE.[in] complete: An optional argument indicating whether the halo updates should be completed before progress resumes. Omitting complete is the same as setting complete to .true.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
- type
The control structure for the MOM_PointAccel module.
Public Members
-
character(len=200) mom_pointaccel::pointaccel_cs::u_trunc_file The complete path to the file in which a column’s worth of u-accelerations are written if u-velocity truncations occur.
-
character(len=200) mom_pointaccel::pointaccel_cs::v_trunc_file The complete path to the file in which a column’s worth of v-accelerations are written if v-velocity truncations occur.
-
integer
u_file¶ The unit number for an opened u-truncation files, or -1 if it has not yet been opened.
-
integer
v_file¶ The unit number for an opened v-truncation files, or -1 if it has not yet been opened.
-
integer
cols_written¶ The number of columns whose output has been written by this PE during the current run.
-
integer
max_writes¶ The maximum number of times any PE can write out a column’s worth of accelerations during a run.
-
type(time_type), pointer mom_pointaccel::pointaccel_cs::time => NULL() A pointer to the ocean model’s clock.
-
type(diag_ctrl), pointer mom_pointaccel::pointaccel_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
real, dimension(:,:,:), pointer mom_pointaccel::pointaccel_cs::u_av => NULL() Time average u-velocity [m s-1].
-
real, dimension(:,:,:), pointer mom_pointaccel::pointaccel_cs::v_av => NULL() Time average velocity [m s-1].
-
real, dimension(:,:,:), pointer mom_pointaccel::pointaccel_cs::u_prev => NULL() Previous u-velocity [m s-1].
-
real, dimension(:,:,:), pointer mom_pointaccel::pointaccel_cs::v_prev => NULL() Previous v-velocity [m s-1].
-
real, dimension(:,:,:), pointer mom_pointaccel::pointaccel_cs::t => NULL() Temperature [degC].
-
real, dimension(:,:,:), pointer mom_pointaccel::pointaccel_cs::s => NULL() Salinity [ppt].
-
real, dimension(:,:,:), pointer mom_pointaccel::pointaccel_cs::u_accel_bt => NULL() Barotropic u-acclerations [m s-2].
-
real, dimension(:,:,:), pointer mom_pointaccel::pointaccel_cs::v_accel_bt => NULL() Barotropic v-acclerations [m s-2].
-
real, dimension(:,:,:), pointer mom_pointaccel::pointaccel_cs::pbce => NULL() pbce times eta gives the baroclinic pressure anomaly in each layer due to free surface height anomalies [m2 s-2 H-1 ~> m s-2 or m4 kg-1 s-2].
-
-
interface
post_data¶ Make a diagnostic available for averaging or output.
Private Functions
-
subroutine
post_data_3d(diag_field_id diag_field_id, field field, diag_cs diag_cs, is_static is_static, mask mask, alt_h alt_h)¶ Make a real 3-d array diagnostic available for averaging or output.
- Parameters
[in] diag_field_id: The id for an output variable returned by a previous call to register_diag_field.[in] field: 3-d array being offered for output or averaging[in] diag_cs: Structure used to regulate diagnostic output[in] is_static: If true, this is a static field that is always offered.[in] mask: If present, use this real array as the data mask.[in] alt_h: An alternate thickness to use for vertically
-
subroutine
post_data_2d(diag_field_id diag_field_id, field field, diag_cs diag_cs, is_static is_static, mask mask)¶ Make a real 2-d array diagnostic available for averaging or output.
- Parameters
[in] diag_field_id: The id for an output variable returned by a previous call to register_diag_field.[in] field: 2-d array being offered for output or averaging[in] diag_cs: Structure used to regulate diagnostic output[in] is_static: If true, this is a static field that is always offered.[in] mask: If present, use this real array as the data mask.
-
subroutine
post_data_1d_k(diag_field_id diag_field_id, field field, diag_cs diag_cs, is_static is_static)¶ Make a real 1-d array diagnostic available for averaging or output.
- Parameters
[in] diag_field_id: The id for an output variable returned by a previous call to register_diag_field.[in] field: 1-d array being offered for output or averaging[in] diag_cs: Structure used to regulate diagnostic output[in] is_static: If true, this is a static field that is always offered.
-
subroutine
post_data_0d(diag_field_id diag_field_id, field field, diag_cs diag_cs, is_static is_static)¶ Make a real scalar diagnostic available for averaging or output.
- Parameters
[in] diag_field_id: The id for an output variable returned by a previous call to register_diag_field.[in] field: real value being offered for output or averaging[in] diag_cs: Structure used to regulate diagnostic output[in] is_static: If true, this is a static field that is always offered.
-
subroutine
- type
Finite volume pressure gradient control structure.
Public Members
-
logical
tides¶ If true, apply tidal momentum forcing.
-
real
rho0¶ The density used in the Boussinesq approximation [kg m-3].
-
real
gfs_scale¶ A scaling of the surface pressure gradients to allow the use of a reduced gravity model [nondim].
-
type(time_type), pointer mom_pressureforce_afv::pressureforce_afv_cs::time A pointer to the ocean model’s clock.
-
type(diag_ctrl), pointer mom_pressureforce_afv::pressureforce_afv_cs::diag A structure that is used to regulate the timing of diagnostic output.
-
logical
usemasswghtinterp¶ Use mass weighting in T/S interpolation.
-
logical
boundary_extrap¶ Indicate whether high-order boundary extrapolation should be used within boundary cells.
-
logical
reconstruct¶ If true, polynomial profiles of T & S will be reconstructed and used in the integrals for the finite volume pressure gradient calculation. The default depends on whether regridding is being used.
-
integer
recon_scheme¶ Order of the polynomial of the reconstruction of T & S for the finite volume pressure gradient calculation. By the default (1) is for a piecewise linear method.
-
integer
id_e_tidal= -1¶ Diagnostic identifier.
-
type(tidal_forcing_cs), pointer mom_pressureforce_afv::pressureforce_afv_cs::tides_csp => NULL() Tides control structure.
-
logical
- type
Finite volume pressure gradient control structure.
Public Members
-
logical
tides¶ If true, apply tidal momentum forcing.
-
real
rho0¶ The density used in the Boussinesq approximation [kg m-3].
-
real
gfs_scale¶ A scaling of the surface pressure gradients to allow the use of a reduced gravity model [nondim].
-
type(time_type), pointer mom_pressureforce_blk_afv::pressureforce_blk_afv_cs::time A pointer to the ocean model’s clock.
-
type(diag_ctrl), pointer mom_pressureforce_blk_afv::pressureforce_blk_afv_cs::diag A structure that is used to regulate the timing of diagnostic output.
-
logical
usemasswghtinterp¶ Use mass weighting in T/S interpolation.
-
logical
boundary_extrap¶ Indicate whether high-order boundary extrapolation should be used within boundary cells.
-
logical
reconstruct¶ If true, polynomial profiles of T & S will be reconstructed and used in the integrals for the finite volume pressure gradient calculation. The default depends on whether regridding is being used.
-
integer
recon_scheme¶ Order of the polynomial of the reconstruction of T & S for the finite volume pressure gradient calculation. By the default (1) is for a piecewise linear method.
-
integer
id_e_tidal= -1¶ Diagnostic identifier.
-
type(tidal_forcing_cs), pointer mom_pressureforce_blk_afv::pressureforce_blk_afv_cs::tides_csp => NULL() Tides control structure.
-
logical
- type
Pressure force control structure.
Public Members
-
logical
analytic_fv_pgf¶ If true, use the analytic finite volume form (Adcroft et al., Ocean Mod. 2008) of the PGF.
-
logical
blocked_afv¶ If true, used the blocked version of the ANALYTIC_FV_PGF code. The value of this parameter should not change answers.
-
type(pressureforce_afv_cs), pointer mom_pressureforce::pressureforce_cs::pressureforce_afv_csp => NULL() Control structure for the analytically integrated finite volume pressure force.
-
type(pressureforce_blk_afv_cs), pointer mom_pressureforce::pressureforce_cs::pressureforce_blk_afv_csp => NULL() Control structure for the analytically integrated finite volume pressure force.
-
type(pressureforce_mont_cs), pointer mom_pressureforce::pressureforce_cs::pressureforce_mont_csp => NULL() Control structure for the Montgomery potential form of pressure force.
-
logical
- type
Control structure for the Montgomery potential form of pressure gradient.
Unnamed Group
-
integer
id_pfu_bc= -1¶ Diagnostic IDs.
-
integer
id_pfv_bc= -1¶ Diagnostic IDs.
-
integer
id_e_tidal= -1¶ Diagnostic IDs.
-
type(tidal_forcing_cs), pointer mom_pressureforce_mont::pressureforce_mont_cs::tides_csp => NULL() The tidal forcing control structure.
Public Members
-
logical
tides¶ If true, apply tidal momentum forcing.
-
real
rho0¶ The density used in the Boussinesq approximation [kg m-3].
-
real
rho_atm¶ The assumed atmospheric density [kg m-3]. By default, Rho_atm is 0.
-
real
gfs_scale¶ Ratio between gravity applied to top interface and the gravitational acceleration of the planet [nondim]. Usually this ratio is 1.
-
type(time_type), pointer mom_pressureforce_mont::pressureforce_mont_cs::time => NULL() A pointer to the ocean model’s clock.
-
type(diag_ctrl), pointer mom_pressureforce_mont::pressureforce_mont_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
real, dimension(:,:,:), pointer mom_pressureforce_mont::pressureforce_mont_cs::pfu_bc => NULL() Accelerations due to pressure.
-
real, dimension(:,:,:), pointer mom_pressureforce_mont::pressureforce_mont_cs::pfv_bc => NULL() gradients deriving from density gradients within layers [m s-2].
-
integer
- type
The control structure for the pseudo-salt tracer.
Public Members
-
type(time_type), pointer pseudo_salt_tracer::pseudo_salt_tracer_cs::time => NULL() A pointer to the ocean model’s clock.
-
type(tracer_registry_type), pointer pseudo_salt_tracer::pseudo_salt_tracer_cs::tr_reg => NULL() A pointer to the MOM tracer registry.
-
real, dimension(:,:,:), pointer pseudo_salt_tracer::pseudo_salt_tracer_cs::ps => NULL() The array of pseudo-salt tracer used in this subroutine [ppt}.
-
real, dimension(:,:,:), pointer pseudo_salt_tracer::pseudo_salt_tracer_cs::diff => NULL() The difference between the pseudo-salt tracer and the real salt [ppt].
-
logical
pseudo_salt_may_reinit= .true.¶ Hard coding since this should not matter.
-
integer
id_psd= -1¶ A diagnostic ID.
-
type(diag_ctrl), pointer pseudo_salt_tracer::pseudo_salt_tracer_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
type(mom_restart_cs), pointer pseudo_salt_tracer::pseudo_salt_tracer_cs::restart_csp => NULL() A pointer to the restart control structure.
-
type(vardesc) pseudo_salt_tracer::pseudo_salt_tracer_cs::tr_desc A description and metadata for the pseudo-salt tracer.
-
- type
A structure with a pointer to a domain2d, to allow for the creation of arrays of pointers.
Private Members
-
type(domain2d), pointer mom_oda_driver_mod::ptr_mpp_domain::mpp_domain => NULL() pointer to an mpp domain2d
-
-
interface
qchksum¶ This is an older interface that has been renamed Bchksum.
Private Functions
-
subroutine
chksum_b_2d(array array, mesg mesg, HI HI, haloshift haloshift, symmetric symmetric, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums a 2d array staggered at corner points.
- Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] mesg: An identifying message[in] haloshift: The width of halos to check (default 0)[in] symmetric: If true, do the checksums on the full symmetric computational domain.[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
chksum_b_3d(array array, mesg mesg, HI HI, haloshift haloshift, symmetric symmetric, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums a 3d array staggered at corner points.
- Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] mesg: An identifying message[in] haloshift: The width of halos to check (default 0)[in] symmetric: If true, do the checksums on the full symmetric computational domain.[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
-
interface
query_initialized¶ Indicate whether a field has been read from a restart file.
Unnamed Group
-
logical function mom_restart::query_initialized::query_initialized_name(name name, CS CS) query_initialized_name determines whether a named field has been successfully read from a restart file yet.
- Parameters
[in] name: The name of the field that is being queriedcs: A pointer to a MOM_restart_CS object (intent in)
-
logical function mom_restart::query_initialized::query_initialized_0d(f_ptr f_ptr, CS CS) Indicate whether the field pointed to by f_ptr has been initialized from a restart file.
- Parameters
[in] f_ptr: A pointer to the field that is being queriedcs: A pointer to a MOM_restart_CS object (intent in)
-
logical function mom_restart::query_initialized::query_initialized_0d_name(f_ptr f_ptr, name name, CS CS) Indicate whether the field pointed to by f_ptr or with the specified variable name has been initialized from a restart file.
- Parameters
[in] f_ptr: A pointer to the field that is being queried[in] name: The name of the field that is being queriedcs: A pointer to a MOM_restart_CS object (intent in)
-
logical function mom_restart::query_initialized::query_initialized_1d(f_ptr f_ptr, CS CS) Indicate whether the field pointed to by f_ptr has been initialized from a restart file.
- Parameters
[in] f_ptr: A pointer to the field that is being queriedcs: A pointer to a MOM_restart_CS object (intent in)
-
logical function mom_restart::query_initialized::query_initialized_1d_name(f_ptr f_ptr, name name, CS CS) Indicate whether the field pointed to by f_ptr or with the specified variable name has been initialized from a restart file.
- Parameters
[in] f_ptr: A pointer to the field that is being queried[in] name: The name of the field that is being queriedcs: A pointer to a MOM_restart_CS object (intent in)
-
logical function mom_restart::query_initialized::query_initialized_2d(f_ptr f_ptr, CS CS) Indicate whether the field pointed to by f_ptr has been initialized from a restart file.
- Parameters
[in] f_ptr: A pointer to the field that is being queriedcs: A pointer to a MOM_restart_CS object (intent in)
-
logical function mom_restart::query_initialized::query_initialized_2d_name(f_ptr f_ptr, name name, CS CS) Indicate whether the field pointed to by f_ptr or with the specified variable name has been initialized from a restart file.
- Parameters
[in] f_ptr: A pointer to the field that is being queried[in] name: The name of the field that is being queriedcs: A pointer to a MOM_restart_CS object (intent in)
-
logical function mom_restart::query_initialized::query_initialized_3d(f_ptr f_ptr, CS CS) Indicate whether the field pointed to by f_ptr has been initialized from a restart file.
- Parameters
[in] f_ptr: A pointer to the field that is being queriedcs: A pointer to a MOM_restart_CS object (intent in)
-
logical function mom_restart::query_initialized::query_initialized_3d_name(f_ptr f_ptr, name name, CS CS) Indicate whether the field pointed to by f_ptr or with the specified variable name has been initialized from a restart file.
- Parameters
[in] f_ptr: A pointer to the field that is being queried[in] name: The name of the field that is being queriedcs: A pointer to a MOM_restart_CS object (intent in)
-
logical function mom_restart::query_initialized::query_initialized_4d(f_ptr f_ptr, CS CS) Indicate whether the field pointed to by f_ptr has been initialized from a restart file.
- Parameters
[in] f_ptr: A pointer to the field that is being queriedcs: A pointer to a MOM_restart_CS object (intent in)
-
logical function mom_restart::query_initialized::query_initialized_4d_name(f_ptr f_ptr, name name, CS CS) Indicate whether the field pointed to by f_ptr or with the specified variable name has been initialized from a restart file.
- Parameters
[in] f_ptr: A pointer to the field that is being queried[in] name: The name of the field that is being queriedcs: A pointer to a MOM_restart_CS object (intent in)
-
-
interface
read_param¶ An overloaded interface to read various types of parameters.
Unnamed Group
-
subroutine
read_param_int(CS CS, varname varname, value value, fail_if_missing fail_if_missing)¶ This subroutine reads the value of an integer model parameter from a parameter file.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file
-
subroutine
read_param_real(CS CS, varname varname, value value, fail_if_missing fail_if_missing, scale scale)¶ This subroutine reads the value of a real model parameter from a parameter file.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file[in] scale: A scaling factor that the parameter is multiplied by before it is returned.
-
subroutine
read_param_logical(CS CS, varname varname, value value, fail_if_missing fail_if_missing)¶ This subroutine reads the value of a logical model parameter from a parameter file.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file
-
subroutine
read_param_char(CS CS, varname varname, value value, fail_if_missing fail_if_missing)¶ This subroutine reads the value of a character string model parameter from a parameter file.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file
-
subroutine
read_param_char_array(CS CS, varname varname, value value, fail_if_missing fail_if_missing)¶ This subroutine reads the values of an array of character string model parameters from a parameter file.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file
-
subroutine
read_param_time(CS CS, varname varname, value value, timeunit timeunit, fail_if_missing fail_if_missing, date_format date_format)¶ This subroutine reads the value of a time_type model parameter from a parameter file.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file[in] timeunit: The number of seconds in a time unit for real-number input.[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file[out] date_format: If present, this indicates whether this parameter was read in a date format, so that it can later be logged in the same format.
-
subroutine
read_param_int_array(CS CS, varname varname, value value, fail_if_missing fail_if_missing)¶ This subroutine reads the values of an array of integer model parameters from a parameter file.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file
-
subroutine
read_param_real_array(CS CS, varname varname, value value, fail_if_missing fail_if_missing, scale scale)¶ This subroutine reads the values of an array of real model parameters from a parameter file.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file[in] scale: A scaling factor that the parameter is multiplied by before it is returned.
-
subroutine
-
interface
register_diag_field_fms¶ A wrapper for register_diag_field_array()
Private Functions
-
integer function mom_diag_manager_wrapper::register_diag_field_fms::register_diag_field_array_fms(module_name module_name, field_name field_name, axes axes, init_time init_time, long_name long_name, units units, missing_value missing_value, range range, mask_variant mask_variant, standard_name standard_name, verbose verbose, do_not_log do_not_log, err_msg err_msg, interp_method interp_method, tile_count tile_count, area area, volume volume) An integer handle for a diagnostic array returned by register_diag_field()
- Parameters
[in] module_name: Name of this module, usually “ocean_model” or “ice_shelf_model”[in] field_name: Name of the diagnostic field[in] axes: Container w/ up to 3 integer handles that indicates axes for this field[in] init_time: Time at which a field is first available?[in] long_name: Long name of a field.[in] units: Units of a field.[in] standard_name: Standardized name associated with a field[in] missing_value: A value that indicates missing values.[in] range: Valid range of a variable (not used in MOM?)[in] mask_variant: If true a logical mask must be provided with post_data calls (not used in MOM?)[in] verbose: If true, FMS is verbose (not used in MOM?)[in] do_not_log: If true, do not log something (not used in MOM?)[out] err_msg: String into which an error message might be placed (not used in MOM?)[in] interp_method: If ‘none’ indicates the field should not be interpolated as a scalar[in] tile_count: no clue (not used in MOM?)[in] area: The FMS id of cell area[in] volume: The FMS id of cell volume
-
integer function mom_diag_manager_wrapper::register_diag_field_fms::register_diag_field_scalar_fms(module_name module_name, field_name field_name, init_time init_time, long_name long_name, units units, missing_value missing_value, range range, mask_variant mask_variant, standard_name standard_name, verbose verbose, do_not_log do_not_log, err_msg err_msg, interp_method interp_method, tile_count tile_count, area area, volume volume) An integer handle for a diagnostic scalar array returned by register_diag_field()
- Parameters
[in] module_name: Name of this module, usually “ocean_model” or “ice_shelf_model”[in] field_name: Name of the diagnostic field[in] init_time: Time at which a field is first available?[in] long_name: Long name of a field.[in] units: Units of a field.[in] standard_name: Standardized name associated with a field[in] missing_value: A value that indicates missing values.[in] range: Valid range of a variable (not used in MOM?)[in] mask_variant: If true a logical mask must be provided with post_data calls (not used in MOM?)[in] verbose: If true, FMS is verbose (not used in MOM?)[in] do_not_log: If true, do not log something (not used in MOM?)[out] err_msg: String into which an error message might be placed (not used in MOM?)[in] interp_method: If ‘none’ indicates the field should not be interpolated as a scalar[in] tile_count: no clue (not used in MOM?)[in] area: The FMS id of cell area (not used for scalars)[in] volume: The FMS id of cell volume (not used for scalars)
-
-
interface
register_restart_field¶ Register fields for restarts.
Unnamed Group
-
subroutine
register_restart_field_ptr4d(f_ptr f_ptr, var_desc var_desc, mandatory mandatory, CS CS)¶ Register a 4-d field for restarts, providing the metadata in a structure.
- Parameters
[in] f_ptr: A pointer to the field to be read or written[in] var_desc: A structure with metadata about this variable[in] mandatory: If true, the run will abort if this field is not successfully read from the restart file.cs: A pointer to a MOM_restart_CS object (intent in/out)
-
subroutine
register_restart_field_4d(f_ptr f_ptr, name name, mandatory mandatory, CS CS, longname longname, units units, hor_grid hor_grid, z_grid z_grid, t_grid t_grid)¶ Register a 4-d field for restarts, providing the metadata as individual arguments.
- Parameters
[in] f_ptr: A pointer to the field to be read or written[in] name: variable name to be used in the restart file[in] mandatory: If true, the run will abort if this field is not successfully read from the restart file.cs: A pointer to a MOM_restart_CS object (intent in/out)[in] longname: variable long name[in] units: variable units[in] hor_grid: variable horizonal staggering, ‘h’ if absent[in] z_grid: variable vertical staggering, ‘L’ if absent[in] t_grid: time description: s, p, or 1, ‘s’ if absent
-
subroutine
register_restart_field_ptr3d(f_ptr f_ptr, var_desc var_desc, mandatory mandatory, CS CS)¶ Register a 3-d field for restarts, providing the metadata in a structure.
- Parameters
[in] f_ptr: A pointer to the field to be read or written[in] var_desc: A structure with metadata about this variable[in] mandatory: If true, the run will abort if this field is not successfully read from the restart file.cs: A pointer to a MOM_restart_CS object (intent in/out)
-
subroutine
register_restart_field_3d(f_ptr f_ptr, name name, mandatory mandatory, CS CS, longname longname, units units, hor_grid hor_grid, z_grid z_grid, t_grid t_grid)¶ Register a 3-d field for restarts, providing the metadata as individual arguments.
- Parameters
[in] f_ptr: A pointer to the field to be read or written[in] name: variable name to be used in the restart file[in] mandatory: If true, the run will abort if this field is not successfully read from the restart file.cs: A pointer to a MOM_restart_CS object (intent in/out)[in] longname: variable long name[in] units: variable units[in] hor_grid: variable horizonal staggering, ‘h’ if absent[in] z_grid: variable vertical staggering, ‘L’ if absent[in] t_grid: time description: s, p, or 1, ‘s’ if absent
-
subroutine
register_restart_field_ptr2d(f_ptr f_ptr, var_desc var_desc, mandatory mandatory, CS CS)¶ Register a 2-d field for restarts, providing the metadata in a structure.
- Parameters
[in] f_ptr: A pointer to the field to be read or written[in] var_desc: A structure with metadata about this variable[in] mandatory: If true, the run will abort if this field is not successfully read from the restart file.cs: A pointer to a MOM_restart_CS object (intent in/out)
-
subroutine
register_restart_field_2d(f_ptr f_ptr, name name, mandatory mandatory, CS CS, longname longname, units units, hor_grid hor_grid, z_grid z_grid, t_grid t_grid)¶ Register a 2-d field for restarts, providing the metadata as individual arguments.
- Parameters
[in] f_ptr: A pointer to the field to be read or written[in] name: variable name to be used in the restart file[in] mandatory: If true, the run will abort if this field is not successfully read from the restart file.cs: A pointer to a MOM_restart_CS object (intent in/out)[in] longname: variable long name[in] units: variable units[in] hor_grid: variable horizonal staggering, ‘h’ if absent[in] z_grid: variable vertical staggering, ‘1’ if absent[in] t_grid: time description: s, p, or 1, ‘s’ if absent
-
subroutine
register_restart_field_ptr1d(f_ptr f_ptr, var_desc var_desc, mandatory mandatory, CS CS)¶ Register a 1-d field for restarts, providing the metadata in a structure.
- Parameters
[in] f_ptr: A pointer to the field to be read or written[in] var_desc: A structure with metadata about this variable[in] mandatory: If true, the run will abort if this field is not successfully read from the restart file.cs: A pointer to a MOM_restart_CS object (intent in/out)
-
subroutine
register_restart_field_1d(f_ptr f_ptr, name name, mandatory mandatory, CS CS, longname longname, units units, hor_grid hor_grid, z_grid z_grid, t_grid t_grid)¶ Register a 1-d field for restarts, providing the metadata as individual arguments.
- Parameters
[in] f_ptr: A pointer to the field to be read or written[in] name: variable name to be used in the restart file[in] mandatory: If true, the run will abort if this field is not successfully read from the restart file.cs: A pointer to a MOM_restart_CS object (intent in/out)[in] longname: variable long name[in] units: variable units[in] hor_grid: variable horizonal staggering, ‘1’ if absent[in] z_grid: variable vertical staggering, ‘L’ if absent[in] t_grid: time description: s, p, or 1, ‘s’ if absent
-
subroutine
register_restart_field_ptr0d(f_ptr f_ptr, var_desc var_desc, mandatory mandatory, CS CS)¶ Register a 0-d field for restarts, providing the metadata in a structure.
- Parameters
[in] f_ptr: A pointer to the field to be read or written[in] var_desc: A structure with metadata about this variable[in] mandatory: If true, the run will abort if this field is not successfully read from the restart file.cs: A pointer to a MOM_restart_CS object (intent in/out)
-
subroutine
register_restart_field_0d(f_ptr f_ptr, name name, mandatory mandatory, CS CS, longname longname, units units, t_grid t_grid)¶ Register a 0-d field for restarts, providing the metadata as individual arguments.
- Parameters
[in] f_ptr: A pointer to the field to be read or written[in] name: variable name to be used in the restart file[in] mandatory: If true, the run will abort if this field is not successfully read from the restart file.cs: A pointer to a MOM_restart_CS object (intent in/out)[in] longname: variable long name[in] units: variable units[in] t_grid: time description: s, p, or 1, ‘s’ if absent
-
subroutine
- type
Regridding control structure.
Public Members
-
real, dimension(:), allocatable mom_regridding::regridding_cs::coordinateresolution This array is set by function setCoordinateResolution() It contains the “resolution” or delta coordinate of the target coorindate. It has the units of the target coordinate, e.g. [Z ~> m] for z*, non-dimensional for sigma, etc.
-
real
coord_scale= 1.0¶ This is a scaling factor that restores coordinateResolution to values in the natural units for output.
-
real, dimension(:), allocatable mom_regridding::regridding_cs::target_density This array is set by function set_target_densities() This array is the nominal coordinate of interfaces and is the running sum of coordinateResolution. i.e. target_density(k+1) = coordinateResolution(k) + coordinateResolution(k) It is only used in “rho” mode.
-
logical
target_density_set= .false.¶ A flag to indicate that the target_density arrays has been filled with data.
-
real, dimension(:), allocatable mom_regridding::regridding_cs::max_interface_depths This array is set by function set_regrid_max_depths() It specifies the maximum depth that every interface is allowed to take [H ~> m or kg m-2].
-
real, dimension(:), allocatable mom_regridding::regridding_cs::max_layer_thickness This array is set by function set_regrid_max_thickness() It specifies the maximum depth that every interface is allowed to take [H ~> m or kg m-2].
-
integer
nk¶ Number of layers/levels in generated grid.
-
integer
regridding_scheme¶ Indicates which grid to use in the vertical (z*, sigma, target interface densities)
-
type(interp_cs_type) mom_regridding::regridding_cs::interp_cs Interpolation control structure.
-
real
min_thickness¶ Minimum thickness allowed when building the new grid through regridding [H ~> m or kg m-2].
-
real
ref_pressure= 2.e7¶ Reference pressure for potential density calculations (Pa)
-
real
old_grid_weight= 0.¶ Weight given to old coordinate when blending between new and old grids [nondim] Used only below depth_of_time_filter_shallow, with a cubic variation from zero to full effect between depth_of_time_filter_shallow and depth_of_time_filter_deep.
-
real
depth_of_time_filter_shallow= 0.¶ Depth above which no time-filtering of grid is applied [H ~> m or kg m-2].
-
real
depth_of_time_filter_deep= 0.¶ Depth below which time-filtering of grid is applied at full effect [H ~> m or kg m-2].
-
real
compressibility_fraction= 0.¶ Fraction (between 0 and 1) of compressibility to add to potential density profiles when interpolating for target grid positions. [nondim].
-
logical
set_maximum_depths= .false.¶ If true, each interface is given a maximum depth based on a rescaling of the indexing of coordinateResolution.
-
real
max_depth_index_scale= 2.0¶ A scaling factor (> 1) of the rate at which the coordinateResolution list is traversed to set the minimum depth of interfaces.
-
logical
integrate_downward_for_e= .true.¶ If true, integrate for interface positions from the top downward. If false, integrate from the bottom upward, as does the rest of the model.
-
type(zlike_cs), pointer mom_regridding::regridding_cs::zlike_cs => null() Control structure for z-like coordinate generator.
-
type(sigma_cs), pointer mom_regridding::regridding_cs::sigma_cs => null() Control structure for sigma coordinate generator.
-
type(rho_cs), pointer mom_regridding::regridding_cs::rho_cs => null() Control structure for rho coordinate generator.
-
type(hycom_cs), pointer mom_regridding::regridding_cs::hycom_cs => null() Control structure for hybrid coordinate generator.
-
type(slight_cs), pointer mom_regridding::regridding_cs::slight_cs => null() Control structure for Slight-coordinate generator.
-
type(adapt_cs), pointer mom_regridding::regridding_cs::adapt_cs => null() Control structure for adaptive coordinate generator.
-
- type
This control structure holds parameters used by the MOM_regularize_layers module.
Unnamed Group
-
integer
id_def_rat_2= -1¶ Diagnostic IDs.
-
integer
id_def_rat_3= -1¶ Diagnostic IDs.
-
integer
id_def_rat_u= -1¶ Diagnostic IDs.
-
integer
id_def_rat_v= -1¶ Diagnostic IDs.
-
integer
id_e1= -1¶ Diagnostic IDs.
-
integer
id_e2= -1¶ Diagnostic IDs.
-
integer
id_e3= -1¶ Diagnostic IDs.
-
integer
id_def_rat_u_1b= -1¶ Diagnostic IDs.
-
integer
id_def_rat_v_1b= -1¶ Diagnostic IDs.
-
integer
id_def_rat_u_2= -1¶ Diagnostic IDs.
-
integer
id_def_rat_u_2b= -1¶ Diagnostic IDs.
-
integer
id_def_rat_v_2= -1¶ Diagnostic IDs.
-
integer
id_def_rat_v_2b= -1¶ Diagnostic IDs.
-
integer
id_def_rat_u_3= -1¶ Diagnostic IDs.
-
integer
id_def_rat_u_3b= -1¶ Diagnostic IDs.
-
integer
id_def_rat_v_3= -1¶ Diagnostic IDs.
-
integer
id_def_rat_v_3b= -1¶ Diagnostic IDs.
Public Members
-
logical
regularize_surface_layers¶ If true, vertically restructure the near-surface layers when they have too much lateral variations to allow for sensible lateral barotropic transports.
-
logical
reg_sfc_detrain¶ If true, allow the buffer layers to detrain into the interior as a part of the restructuring when regularize_surface_layers is true.
-
real
h_def_tol1¶ The value of the relative thickness deficit at which to start modifying the structure, 0.5 by default (or a thickness ratio of 5.83).
-
real
h_def_tol2¶ The value of the relative thickness deficit at which to the structure modification is in full force, now 20% of the way from h_def_tol1 to 1.
-
real
h_def_tol3¶ The value of the relative thickness deficit at which to start detrainment from the buffer layers to the interior, now 30% of the way from h_def_tol1 to 1.
-
real
h_def_tol4¶ The value of the relative thickness deficit at which to do detrainment from the buffer layers to the interior at full force, now 50% of the way from h_def_tol1 to 1.
-
real
hmix_min¶ The minimum mixed layer thickness [H ~> m or kg m-2].
-
type(time_type), pointer mom_regularize_layers::regularize_layers_cs::time => NULL() A pointer to the ocean model’s clock.
-
type(diag_ctrl), pointer mom_regularize_layers::regularize_layers_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
logical
debug¶ If true, do more thorough checks for debugging purposes.
-
integer
id_def_rat= -1¶ A diagnostic ID.
-
logical
allow_clocks_in_omp_loops¶ If true, clocks can be called from inside loops that can be threaded. To run with multiple threads, set to False.
-
integer
- type
Container for remapping parameters.
Public Members
-
integer
remapping_scheme= -911¶ Determines which reconstruction to use.
-
integer
degree= 0¶ Degree of polynomial reconstruction.
-
logical
boundary_extrapolation= .true.¶ If true, extrapolate boundaries.
-
logical
check_reconstruction= .false.¶ If true, reconstructions are checked for consistency.
-
logical
check_remapping= .false.¶ If true, the result of remapping are checked for conservation and bounds.
-
logical
force_bounds_in_subcell= .false.¶ If true, the intermediate values used in remapping are forced to be bounded.
-
integer
-
interface
reproducing_sum¶ Find an accurate and order-invariant sum of distributed 2d or 3d fields.
Private Functions
-
real function mom_coms::reproducing_sum::reproducing_sum_2d(array array, isr isr, ier ier, jsr jsr, jer jer, EFP_sum EFP_sum, reproducing reproducing, overflow_check overflow_check, err err) This subroutine uses a conversion to an integer representation of real numbers to give an order-invariant sum of distributed 2-D arrays that reproduces across domain decomposition. This technique is described in Hallberg & Adcroft, 2014, Parallel Computing, doi:10.1016/j.parco.2014.04.007.
- Return
Result
- Parameters
[in] array: The array to be summed[in] isr: The starting i-index of the sum, noting that the array indices starts at 1[in] ier: The ending i-index of the sum, noting that the array indices starts at 1[in] jsr: The starting j-index of the sum, noting that the array indices starts at 1[in] jer: The ending j-index of the sum, noting that the array indices starts at 1[out] efp_sum: The result in extended fixed point format[in] reproducing: If present and false, do the sum using the naive non-reproducing approach[in] overflow_check: If present and false, disable checking for overflows in incremental results. This can speed up calculations if the number of values being summed is small enough[out] err: If present, return an error code instead of triggering any fatal errors directly from this routine.
-
real function mom_coms::reproducing_sum::reproducing_sum_3d(array array, isr isr, ier ier, jsr jsr, jer jer, sums sums, EFP_sum EFP_sum, err err) This subroutine uses a conversion to an integer representation of real numbers to give an order-invariant sum of distributed 3-D arrays that reproduces across domain decomposition. This technique is described in Hallberg & Adcroft, 2014, Parallel Computing, doi:10.1016/j.parco.2014.04.007.
- Return
Result
- Parameters
[in] array: The array to be summed[in] isr: The starting i-index of the sum, noting that the array indices starts at 1[in] ier: The ending i-index of the sum, noting that the array indices starts at 1[in] jsr: The starting j-index of the sum, noting that the array indices starts at 1[in] jer: The ending j-index of the sum, noting that the array indices starts at 1[out] sums: The sums by vertical layer[out] efp_sum: The result in extended fixed point format[out] err: If present, return an error code instead of triggering any fatal errors directly from this routine.
-
- type
tracer control structure
Public Members
-
logical
coupled_tracers= .false.¶ These tracers are not offered to the coupler.
-
character(len = 200) rgc_tracer::rgc_tracer_cs::tracer_ic_file The full path to the IC file, or ” ” to initialize internally.
-
type(time_type), pointer rgc_tracer::rgc_tracer_cs::time A pointer to the ocean model’s clock.
-
type(tracer_registry_type), pointer rgc_tracer::rgc_tracer_cs::tr_reg => NULL() A pointer to the tracer registry.
-
real, dimension(:,:,:,:), pointer rgc_tracer::rgc_tracer_cs::tr => NULL() The array of tracers used in this package.
-
real, dimension(:,:,:,:), pointer rgc_tracer::rgc_tracer_cs::tr_aux => NULL() The masked tracer concentration.
-
real, dimension(ntr) rgc_tracer::rgc_tracer_cs::land_val = -1.0 The value of tr used where land is masked out.
-
real
lenlat¶ the latitudinal or y-direction length of the domain.
-
real
lenlon¶ the longitudinal or x-direction length of the domain.
-
real
csl¶ The length of the continental shelf (x dir, km)
-
real
lensponge¶ the length of the sponge layer.
-
logical
mask_tracers¶ If true, tracers are masked out in massless layers.
-
logical
use_sponge¶ If true, sponges may be applied somewhere in the domain.
-
type(diag_ctrl), pointer rgc_tracer::rgc_tracer_cs::diag A structure that is used to regulate the timing of diagnostic output.
-
type(vardesc), dimension(ntr) rgc_tracer::rgc_tracer_cs::tr_desc Descriptions and metadata for the tracers.
-
logical
- type
Control structure containing required parameters for the rho coordinate.
Public Members
-
integer
nk¶ Number of layers.
-
real
min_thickness= 0.¶ Minimum thickness allowed for layers, often in [H ~> m or kg m-2].
-
real
ref_pressure¶ Reference pressure for density calculations [Pa].
-
logical
integrate_downward_for_e= .false.¶ If true, integrate for interface positions from the top downward. If false, integrate from the bottom upward, as does the rest of the model.
-
real, dimension(:), allocatable coord_rho::rho_cs::target_density Nominal density of interfaces [kg m-3].
-
type(interp_cs_type) coord_rho::rho_cs::interp_cs Interpolation control structure.
-
integer
-
interface
safe_alloc_alloc¶ Allocate a 2-d or 3-d allocatable array.
Private Functions
-
subroutine
safe_alloc_allocatable_3d(ptr ptr, is is, ie ie, js js, je je, nk nk)¶ Allocate a 3-d allocatable array based on its index starting and ending values and k-index size.
- Parameters
ptr: An allocatable array to allocate[in] is: The start index to allocate for the 1st dimension[in] ie: The end index to allocate for the 1st dimension[in] js: The start index to allocate for the 2nd dimension[in] je: The end index to allocate for the 2nd dimension[in] nk: The size to allocate for the 3rd dimension
-
subroutine
safe_alloc_allocatable_2d(ptr ptr, is is, ie ie, js js, je je)¶ Allocate a 2-d allocatable array based on its index starting and ending values.
- Parameters
ptr: An allocatable array to allocate[in] is: The start index to allocate for the 1st dimension[in] ie: The end index to allocate for the 1st dimension[in] js: The start index to allocate for the 2nd dimension[in] je: The end index to allocate for the 2nd dimension
-
subroutine
safe_alloc_allocatable_3d_6arg(ptr ptr, is is, ie ie, js js, je je, ks ks, ke ke)¶ Allocate a 3-d allocatable array based on its 6 index starting and ending values.
- Parameters
ptr: An allocatable array to allocate[in] is: The start index to allocate for the 1st dimension[in] ie: The end index to allocate for the 1st dimension[in] js: The start index to allocate for the 2nd dimension[in] je: The end index to allocate for the 2nd dimension[in] ks: The start index to allocate for the 3rd dimension[in] ke: The end index to allocate for the 3rd dimension
-
subroutine
-
interface
safe_alloc_ptr¶ Allocate a pointer to a 1-d, 2-d or 3-d array.
Private Functions
-
subroutine
safe_alloc_ptr_3d_3arg(ptr ptr, ni ni, nj nj, nk nk)¶ Allocate a pointer to a 3-d array based on its dimension sizes.
- Parameters
ptr: A pointer to allocate[in] ni: The size of the 1st dimension of the array[in] nj: The size of the 2nd dimension of the array[in] nk: The size of the 3rd dimension of the array
-
subroutine
safe_alloc_ptr_3d_6arg(ptr ptr, is is, ie ie, js js, je je, ks ks, ke ke)¶ Allocate a pointer to a 3-d array based on its index starting and ending values.
- Parameters
ptr: A pointer to allocate[in] is: The start index to allocate for the 1st dimension[in] ie: The end index to allocate for the 1st dimension[in] js: The start index to allocate for the 2nd dimension[in] je: The end index to allocate for the 2nd dimension[in] ks: The start index to allocate for the 3rd dimension[in] ke: The end index to allocate for the 3rd dimension
-
subroutine
safe_alloc_ptr_2d_2arg(ptr ptr, ni ni, nj nj)¶ Allocate a pointer to a 2-d array based on its dimension sizes.
- Parameters
ptr: A pointer to allocate[in] ni: The size of the 1st dimension of the array[in] nj: The size of the 2nd dimension of the array
-
subroutine
safe_alloc_ptr_3d(ptr ptr, is is, ie ie, js js, je je, nk nk)¶ Allocate a pointer to a 3-d array based on its index starting and ending values.
- Parameters
ptr: A pointer to allocate[in] is: The start index to allocate for the 1st dimension[in] ie: The end index to allocate for the 1st dimension[in] js: The start index to allocate for the 2nd dimension[in] je: The end index to allocate for the 2nd dimension[in] nk: The size to allocate for the 3rd dimension
-
subroutine
safe_alloc_ptr_2d(ptr ptr, is is, ie ie, js js, je je)¶ Allocate a pointer to a 2-d array based on its index starting and ending values.
- Parameters
ptr: A pointer to allocate[in] is: The start index to allocate for the 1st dimension[in] ie: The end index to allocate for the 1st dimension[in] js: The start index to allocate for the 2nd dimension[in] je: The end index to allocate for the 2nd dimension
-
subroutine
- type
Container for surface forcing parameters.
Public Members
-
real, public scm_cvmix_tests::scm_cvmix_tests_cs::rho0 reference density copied for easy passing [kg m-3]
Private Members
-
logical
usewindstress¶ True to use wind stress.
-
logical
useheatflux¶ True to use heat flux.
-
logical
useevaporation¶ True to use evaporation.
-
logical
usediurnalsw¶ True to use diurnal sw radiation.
-
real
tau_x¶ (Constant) Wind stress, X [Pa]
-
real
tau_y¶ (Constant) Wind stress, Y [Pa]
-
real
surf_hf¶ (Constant) Heat flux [m degC s-1]
-
real
surf_evap¶ (Constant) Evaporation rate [m s-1]
-
real
max_sw¶ maximum of diurnal sw radiation [m degC s-1]
-
- type
Registry type for tracers on segments.
Public Members
-
integer
ntseg= 0¶ number of registered tracer segments
-
type(obc_segment_tracer_type), dimension( 50 ) mom_open_boundary::segment_tracer_registry_type::tr array of registered tracers
-
logical
locked= .false.¶ New tracers may be registered if locked=.false. When locked=.true.,no more tracers can be registered. Not sure who should lock it or when…
-
integer
- type
This control structure contains parameters for MOM_set_diffusivity.
Unnamed Group
-
integer
id_maxtke= -1¶ Diagnostic IDs.
-
integer
id_tke_to_kd= -1¶ Diagnostic IDs.
-
integer
id_kd_user= -1¶ Diagnostic IDs.
-
integer
id_kd_layer= -1¶ Diagnostic IDs.
-
integer
id_kd_bbl= -1¶ Diagnostic IDs.
-
integer
id_n2= -1¶ Diagnostic IDs.
-
integer
id_kd_work= -1¶ Diagnostic IDs.
-
integer
id_kt_extra= -1¶ Diagnostic IDs.
-
integer
id_ks_extra= -1¶ Diagnostic IDs.
Public Members
-
logical
debug¶ If true, write verbose checksums for debugging.
-
logical
bulkmixedlayer¶ If true, a refined bulk mixed layer is used with GVnk_rho_varies variable density mixed & buffer layers.
-
real
fluxri_max¶ The flux Richardson number where the stratification is large enough that N2 > omega2. The full expression for the Flux Richardson number is usually FLUX_RI_MAX*N2/(N2+OMEGA2). The default is 0.2.
-
logical
bottomdraglaw¶ If true, the bottom stress is calculated with a drag law c_drag*|u|*u.
-
logical
bbl_mixing_as_max¶ If true, take the maximum of the diffusivity from the BBL mixing and the other diffusivities. Otherwise, diffusivities from the BBL_mixing is added.
-
logical
use_lotw_bbl_diffusivity¶ If true, use simpler/less precise, BBL diffusivity.
-
logical
lotw_bbl_use_omega¶ If true, use simpler/less precise, BBL diffusivity.
-
real
bbl_effic¶ efficiency with which the energy extracted by bottom drag drives BBL diffusion [nondim]
-
real
cdrag¶ quadratic drag coefficient [nondim]
-
real
imax_decay¶ inverse of a maximum decay scale for bottom-drag driven turbulence [Z-1 ~> m-1].
-
real
kv¶ The interior vertical viscosity [Z2 T-1 ~> m2 s-1].
-
real
kd¶ interior diapycnal diffusivity [Z2 T-1 ~> m2 s-1].
-
real
kd_min¶ minimum diapycnal diffusivity [Z2 T-1 ~> m2 s-1].
-
real
kd_max¶ maximum increment for diapycnal diffusivity [Z2 T-1 ~> m2 s-1]. Set to a negative value to have no limit.
-
real
kd_add¶ uniform diffusivity added everywhere without filtering or scaling [Z2 T-1 ~> m2 s-1].
-
real
kd_smooth¶ Vertical diffusivity used to interpolate more sensible values of T & S into thin layers [Z2 T-1 ~> m2 s-1].
-
type(diag_ctrl), pointer mom_set_diffusivity::set_diffusivity_cs::diag => NULL() structure to regulate diagnostic output timing
-
logical
limit_dissipation¶ If enabled, dissipation is limited to be larger than the following:
-
real
dissip_min¶ Minimum dissipation [kg Z2 m-3 T-3 ~> W m-3].
-
real
dissip_n0¶ Coefficient a in minimum dissipation = a+b*N [kg Z2 m-3 T-3 ~> W m-3].
-
real
dissip_n1¶ Coefficient b in minimum dissipation = a+b*N [kg Z2 m-3 T-2 ~> J m-3].
-
real
dissip_n2¶ Coefficient c in minimum dissipation = c*N2 [kg Z2 m-3 T-1 ~> J s m-3].
-
real
dissip_kd_min¶ Minimum Kd [Z2 T-1 ~> m2 s-1], with dissipation Rho0*Kd_min*N^2.
-
real
tke_itide_max¶ maximum internal tide conversion [W m-2] available to mix above the BBL
-
real
omega¶ Earth’s rotation frequency [T-1 ~> s-1].
-
logical
ml_radiation¶ allow a fraction of TKE available from wind work to penetrate below mixed layer base with a vertical decay scale determined by the minimum of (1) The depth of the mixed layer, or (2) An Ekman length scale. Energy available to drive mixing below the mixed layer is given by E = ML_RAD_COEFF*MSTAR*USTAR**3. Optionally, if ML_rad_TKE_decay is true, this is further reduced by a factor of exp(-h_ML*Idecay_len_TkE), where Idecay_len_TKE is calculated the same way as in the mixed layer code. The diapycnal diffusivity is KD(k) = E/(N2(k)+OMEGA2), where N2 is the squared buoyancy frequency [s-2] and OMEGA2 is the rotation rate of the earth squared.
-
real
ml_rad_kd_max¶ Maximum diapycnal diffusivity due to turbulence radiated from the base of the mixed layer [Z2 T-1 ~> m2 s-1].
-
real
ml_rad_efold_coeff¶ non-dim coefficient to scale penetration depth
-
real
ml_rad_coeff¶ coefficient, which scales MSTAR*USTAR^3 to obtain energy available for mixing below mixed layer base [nondim]
-
logical
ml_rad_bug¶ If true use code with a bug that reduces the energy available in the transition layer by a factor of the inverse of the energy deposition lenthscale (in m).
-
logical
ml_rad_tke_decay¶ If true, apply same exponential decay to ML_rad as applied to the other surface sources of TKE in the mixed layer code.
-
real
ustar_min¶ A minimum value of ustar to avoid numerical problems [Z T-1 ~> m s-1]. If the value is small enough, this parameter should not affect the solution.
-
real
tke_decay¶ ratio of natural Ekman depth to TKE decay scale [nondim]
-
real
mstar¶ ratio of friction velocity cubed to TKE input to the mixed layer [nondim]
-
logical
ml_use_omega¶ If true, use absolute rotation rate instead of the vertical component of rotation when setting the decay scale for mixed layer turbulence.
-
real
ml_omega_frac¶ When setting the decay scale for turbulence, use this fraction of the absolute rotation rate blended with the local value of f, as f^2 ~= (1-of)*f^2 + of*4*omega^2.
-
logical
user_change_diff¶ If true, call user-defined code to change diffusivity.
-
logical
usekappashear¶ If true, use the kappa_shear module to find the shear-driven diapycnal diffusivity.
-
logical
vertex_shear¶ If true, do the calculations of the shear-driven mixing at the cell vertices (i.e., the vorticity points).
-
logical
use_cvmix_shear¶ If true, use one of the CVMix modules to find shear-driven diapycnal diffusivity.
-
logical
double_diffusion¶ If true, enable double-diffusive mixing using an old method.
-
logical
use_cvmix_ddiff¶ If true, enable double-diffusive mixing via CVMix.
-
logical
simple_tke_to_kd¶ If true, uses a simple estimate of Kd/TKE that does not rely on a layer-formulation.
-
real
max_rrho_salt_fingers¶ max density ratio for salt fingering
-
real
max_salt_diff_salt_fingers¶ max salt diffusivity for salt fingers [Z2 T-1 ~> m2 s-1]
-
real
kv_molecular¶ molecular visc for double diff convect [Z2 T-1 ~> m2 s-1]
-
logical
answers_2018¶ If true, use the order of arithmetic and expressions that recover the answers from the end of 2018. Otherwise, use updated and more robust forms of the same expressions.
-
character(len=200) mom_set_diffusivity::set_diffusivity_cs::inputdir The directory in which input files are found.
-
type(user_change_diff_cs), pointer mom_set_diffusivity::set_diffusivity_cs::user_change_diff_csp => NULL() Control structure for a child module.
-
type(kappa_shear_cs), pointer mom_set_diffusivity::set_diffusivity_cs::kappashear_csp => NULL() Control structure for a child module.
-
type(cvmix_shear_cs), pointer mom_set_diffusivity::set_diffusivity_cs::cvmix_shear_csp => NULL() Control structure for a child module.
-
type(cvmix_ddiff_cs), pointer mom_set_diffusivity::set_diffusivity_cs::cvmix_ddiff_csp => NULL() Control structure for a child module.
-
type(bkgnd_mixing_cs), pointer mom_set_diffusivity::set_diffusivity_cs::bkgnd_mixing_csp => NULL() Control structure for a child module.
-
type(int_tide_cs), pointer mom_set_diffusivity::set_diffusivity_cs::int_tide_csp => NULL() Control structure for a child module.
-
type(tidal_mixing_cs), pointer mom_set_diffusivity::set_diffusivity_cs::tm_csp => NULL() Control structure for a child module.
-
integer
-
interface
set_up_ale_sponge_field¶ Store the reference profile at h points for a variable.
Private Functions
-
subroutine
set_up_ale_sponge_field_fixed(sp_val sp_val, G G, f_ptr f_ptr, CS CS)¶ This subroutine stores the reference profile at h points for the variable whose address is given by f_ptr.
- Parameters
[in] g: Grid structurecs: ALE sponge control structure (in/out).[in] sp_val: Field to be used in the sponge, it has arbritary number of layers.[in] f_ptr: Pointer to the field to be damped
-
subroutine
set_up_ale_sponge_field_varying(filename filename, fieldname fieldname, Time Time, G G, GV GV, f_ptr f_ptr, CS CS)¶ This subroutine stores the reference profile at h points for the variable whose address is given by filename and fieldname.
- Parameters
[in] filename: The name of the file with the time varying field data[in] fieldname: The name of the field in the file with the time varying field data[in] time: The current model time[in] g: Grid structure (in).[in] gv: ocean vertical grid structure[in] f_ptr: Pointer to the field to be damped (in).cs: Sponge control structure (in/out).
-
subroutine
-
interface
set_up_ale_sponge_vel_field¶ This subroutine stores the reference profile at u and v points for a vector.
Private Functions
-
subroutine
set_up_ale_sponge_vel_field_fixed(u_val u_val, v_val v_val, G G, u_ptr u_ptr, v_ptr v_ptr, CS CS)¶ This subroutine stores the reference profile at u and v points for the variable whose address is given by u_ptr and v_ptr.
- Parameters
[in] g: Grid structure (in).cs: Sponge structure (in/out).[in] u_val: u field to be used in the sponge, it has arbritary number of layers.[in] v_val: v field to be used in the sponge, it has arbritary number of layers.[in] u_ptr: u pointer to the field to be damped[in] v_ptr: v pointer to the field to be damped
-
subroutine
set_up_ale_sponge_vel_field_varying(filename_u filename_u, fieldname_u fieldname_u, filename_v filename_v, fieldname_v fieldname_v, Time Time, G G, US US, CS CS, u_ptr u_ptr, v_ptr v_ptr)¶ This subroutine stores the reference profile at uand v points for the variable whose address is given by u_ptr and v_ptr.
- Parameters
[in] filename_u: File name for u field[in] fieldname_u: Name of u variable in file[in] filename_v: File name for v field[in] fieldname_v: Name of v variable in file[in] time: Model time[inout] g: Ocean grid (in)[in] us: A dimensional unit scaling typecs: Sponge structure (in/out).[in] u_ptr: u pointer to the field to be damped (in).[in] v_ptr: v pointer to the field to be damped (in).
-
subroutine
- type
Control structure for MOM_set_visc.
Unnamed Group
-
integer
id_bbl_thick_u= -1¶ Diagnostics handles.
-
integer
id_kv_bbl_u= -1¶ Diagnostics handles.
-
integer
id_bbl_thick_v= -1¶ Diagnostics handles.
-
integer
id_kv_bbl_v= -1¶ Diagnostics handles.
-
integer
id_ray_u= -1¶ Diagnostics handles.
-
integer
id_ray_v= -1¶ Diagnostics handles.
-
integer
id_nkml_visc_u= -1¶ Diagnostics handles.
-
integer
id_nkml_visc_v= -1¶ Diagnostics handles.
Public Members
-
real
hbbl¶ The static bottom boundary layer thickness [H ~> m or kg m-2].
-
real
cdrag¶ The quadratic drag coefficient.
-
real
c_smag¶ The Laplacian Smagorinsky coefficient for calculating the drag in channels.
-
real
drag_bg_vel¶ An assumed unresolved background velocity for calculating the bottom drag [m s-1].
-
real
bbl_thick_min¶ The minimum bottom boundary layer thickness [H ~> m or kg m-2]. This might be Kv / (cdrag * drag_bg_vel) to give Kv as the minimum near-bottom viscosity.
-
real
htbl_shelf¶ A nominal thickness of the surface boundary layer for use in calculating the near-surface velocity [H ~> m or kg m-2].
-
real
htbl_shelf_min¶ The minimum surface boundary layer thickness [H ~> m or kg m-2].
-
real
kv_bbl_min¶ The minimum viscosity in the bottom boundary layer [Z2 T-1 ~> m2 s-1].
-
real
kv_tbl_min¶ The minimum viscosity in the top boundary layer [Z2 T-1 ~> m2 s-1].
-
logical
bottomdraglaw¶ If true, the bottom stress is calculated with a drag law c_drag*|u|*u. The velocity magnitude may be an assumed value or it may be based on the actual velocity in the bottommost HBBL, depending on whether linear_drag is true.
-
logical
bbl_use_eos¶ If true, use the equation of state in determining the properties of the bottom boundary layer.
-
logical
linear_drag¶ If true, the drag law is cdrag*DRAG_BG_VEL*u.
-
logical
channel_drag¶ If true, the drag is exerted directly on each layer according to what fraction of the bottom they overlie.
-
logical
rino_mix¶ If true, use Richardson number dependent mixing.
-
logical
dynamic_viscous_ml¶ If true, use a bulk Richardson number criterion to determine the mixed layer thickness for viscosity.
-
real
bulk_ri_ml¶ The bulk mixed layer used to determine the thickness of the viscous mixed layer. Nondim.
-
real
omega¶ The Earth’s rotation rate [T-1 ~> s-1].
-
real
ustar_min¶ A minimum value of ustar to avoid numerical problems [Z T-1 ~> m s-1]. If the value is small enough, this should not affect the solution.
-
real
tke_decay¶ The ratio of the natural Ekman depth to the TKE decay scale, nondimensional.
-
real
omega_frac¶ When setting the decay scale for turbulence, use this fraction of the absolute rotation rate blended with the local value of f, as sqrt((1-of)*f^2 + of*4*omega^2).
-
logical
answers_2018¶ If true, use the order of arithmetic and expressions that recover the answers from the end of 2018. Otherwise, use updated and more robust forms of the same expressions.
-
logical
debug¶ If true, write verbose checksums for debugging purposes.
-
type(ocean_obc_type), pointer mom_set_visc::set_visc_cs::obc => NULL() Open boundaries control structure.
-
type(diag_ctrl), pointer mom_set_visc::set_visc_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
integer
- type
Control structure for shelfwave open boundaries.
- type
Control structure containing required parameters for the sigma coordinate.
- type
Control structure containing required parameters for the SLight coordinate.
Public Members
-
integer
nk¶ Number of layers/levels.
-
real
min_thickness¶ Minimum thickness allowed when building the new grid through regridding [H ~> m or kg m-2].
-
real
ref_pressure¶ Reference pressure for potential density calculations [Pa].
-
real
compressibility_fraction¶ Fraction (between 0 and 1) of compressibility to add to potential density profiles when interpolating for target grid positions. [nondim].
-
real
rho_ml_avg_depth¶ Depth over which to average to determine the mixed layer potential density [H ~> m or kg m-2].
-
real
nlay_ml_offset¶ Number of layers to offset the mixed layer density to find resolved stratification [nondim].
-
integer
nz_fixed_surface= 2¶ The number of fixed-thickness layers at the top of the model.
-
real
dz_ml_min¶ The fixed resolution in the topmost SLight_nkml_min layers [H ~> m or kg m-2].
-
logical
fix_haloclines= .false.¶ If true, detect regions with much weaker stratification in the coordinate than based on in-situ density, and use a stretched coordinate there.
-
real
halocline_filter_length¶ A length scale over which to filter T & S when looking for spuriously unstable water mass profiles [H ~> m or kg m-2].
-
real
halocline_strat_tol¶ A value of the stratification ratio that defines a problematic halocline region [nondim].
-
real, dimension(:), allocatable coord_slight::slight_cs::target_density Nominal density of interfaces [kg m-3].
-
real, dimension(:), allocatable coord_slight::slight_cs::max_interface_depths Maximum depths of interfaces [H ~> m or kg m-2].
-
real, dimension(:), allocatable coord_slight::slight_cs::max_layer_thickness Maximum thicknesses of layers [H ~> m or kg m-2].
-
type(interp_cs_type) coord_slight::slight_cs::interp_cs Interpolation control structure.
-
integer
- type
This control structure holds memory and parameters for the MOM_sponge module.
Public Members
-
logical
bulkmixedlayer¶ If true, a refined bulk mixed layer is used with nkml sublayers and nkbl buffer layer.
-
integer
nz¶ The total number of layers.
-
integer
isc¶ The starting i-index of the computational domain at h.
-
integer
iec¶ The ending i-index of the computational domain at h.
-
integer
jsc¶ The starting j-index of the computational domain at h.
-
integer
jec¶ The ending j-index of the computational domain at h.
-
integer
isd¶ The starting i-index of the data domain at h.
-
integer
ied¶ The ending i-index of the data domain at h.
-
integer
jsd¶ The starting j-index of the data domain at h.
-
integer
jed¶ The ending j-index of the data domain at h.
-
integer
num_col¶ The number of sponge points within the computational domain.
-
integer
fldno= 0¶ The number of fields which have already been registered by calls to set_up_sponge_field.
-
integer, dimension(:), pointer mom_sponge::sponge_cs::col_i => NULL() Array of the i-indicies of each of the columns being damped.
-
integer, dimension(:), pointer mom_sponge::sponge_cs::col_j => NULL() Array of the j-indicies of each of the columns being damped.
-
real, dimension(:), pointer mom_sponge::sponge_cs::iresttime_col => NULL() The inverse restoring time of each column.
-
real, dimension(:), pointer mom_sponge::sponge_cs::rcv_ml_ref => NULL() The value toward which the mixed layer coordinate-density is being damped [kg m-3].
-
real, dimension(:,:), pointer mom_sponge::sponge_cs::ref_eta => NULL() The value toward which the interface heights are being damped [Z ~> m].
-
type(p3d), dimension( 50 ) mom_sponge::sponge_cs::var Pointers to the fields that are being damped.
-
type(p2d), dimension( 50 ) mom_sponge::sponge_cs::ref_val The values to which the fields are damped.
-
logical
do_i_mean_sponge¶ If true, apply sponges to the i-mean fields.
-
real, dimension(:), pointer mom_sponge::sponge_cs::iresttime_im => NULL() The inverse restoring time of each row for i-mean sponges.
-
real, dimension(:), pointer mom_sponge::sponge_cs::rcv_ml_ref_im => NULL() mixed layer coordinate-density is being damped [kg m-3].
-
real, dimension(:,:), pointer mom_sponge::sponge_cs::ref_eta_im => NULL() The value toward which the i-mean interface heights are being damped [Z ~> m].
-
type(p2d), dimension( 50 ) mom_sponge::sponge_cs::ref_val_im The values toward which the i-means of fields are damped.
-
type(diag_ctrl), pointer mom_sponge::sponge_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
integer
id_w_sponge= -1¶ A diagnostic ID.
-
logical
-
interface
state_dependent¶ Returns true if the coordinate is dependent on the state density, returns false otherwise.
Public Functions
-
logical function regrid_consts::state_dependent::state_dependent_char(string string) Returns true if the coordinate is dependent on the state density, returns false otherwise.
- Parameters
[in] string: String to indicate coordinate mode
-
logical function regrid_consts::state_dependent::state_dependent_int(mode mode) Returns true if the coordinate is dependent on the state density, returns false otherwise.
- Parameters
[in] mode: Coordinate mode
-
- type
A type for storing statistica about a variable.
- type
The control structure for the MOM_sum_output module.
Public Members
-
type(depth_list), dimension(:), pointer mom_sum_output::sum_output_cs::dl => NULL() The sorted depth list.
-
integer
list_size¶ length of sorting vector <= niglobal*njglobal
-
integer, dimension(:), allocatable mom_sum_output::sum_output_cs::lh This saves the entry in DL with a volume just less than the volume of fluid below the interface.
-
logical
do_ape_calc¶ If true, calculate the available potential energy of the interfaces. Disabling this reduces the memory footprint of high-PE-count models dramatically.
-
logical
read_depth_list¶ Read the depth list from a file if it exists and write it if it doesn’t.
-
character(len=200) mom_sum_output::sum_output_cs::depth_list_file The name of the depth list file.
-
real
d_list_min_inc¶ The minimum increment [Z ~> m], between the depths of the entries in the depth-list file, 0 by default.
-
logical
require_depth_list_chksum¶ Require matching checksums in Depth_list.nc when reading the file.
-
logical
update_depth_list_chksum¶ Automatically update the Depth_list.nc file if the checksums are missing or do not match current values.
-
logical
use_temperature¶ If true, temperature and salinity are state variables.
-
real
fresh_water_input¶ The total mass of fresh water added by surface fluxes since the last time that write_energy was called [kg].
-
real
mass_prev¶ The total ocean mass the last time that write_energy was called [kg].
-
real
salt_prev¶ The total amount of salt in the ocean the last time that write_energy was called [ppt kg].
-
real
net_salt_input¶ The total salt added by surface fluxes since the last time that write_energy was called [ppt kg].
-
real
heat_prev¶ The total amount of heat in the ocean the last time that write_energy was called [J].
-
real
net_heat_input¶ The total heat added by surface fluxes since the last the last time that write_energy was called [J].
-
type(efp_type) mom_sum_output::sum_output_cs::fresh_water_in_efp An extended fixed point version of fresh_water_input.
-
type(efp_type) mom_sum_output::sum_output_cs::net_salt_in_efp An extended fixed point version of net_salt_input.
-
type(efp_type) mom_sum_output::sum_output_cs::net_heat_in_efp An extended fixed point version of net_heat_input.
-
type(efp_type) mom_sum_output::sum_output_cs::heat_prev_efp An extended fixed point version of heat_prev.
-
type(efp_type) mom_sum_output::sum_output_cs::salt_prev_efp An extended fixed point version of salt_prev.
-
type(efp_type) mom_sum_output::sum_output_cs::mass_prev_efp An extended fixed point version of mass_prev.
-
real
dt¶ The baroclinic dynamics time step [s].
-
type(time_type) mom_sum_output::sum_output_cs::energysavedays The interval between writing the energies and other integral quantities of the run.
-
type(time_type) mom_sum_output::sum_output_cs::energysavedays_geometric The starting interval for computing a geometric progression of time deltas between calls to write_energy. This interval will increase by a factor of 2. after each call to write_energy.
-
logical
energysave_geometric¶ Logical to control whether calls to write_energy should follow a geometric progression.
-
type(time_type) mom_sum_output::sum_output_cs::write_energy_time The next time to write to the energy file.
-
type(time_type) mom_sum_output::sum_output_cs::geometric_end_time Time at which to stop the geometric progression of calls to write_energy and revert to the standard energysavedays interval.
-
real
timeunit¶ The length of the units for the time axis [s].
-
logical
date_stamped_output¶ If true, use dates (not times) in messages to stdout.
-
type(time_type) mom_sum_output::sum_output_cs::start_time The start time of the simulation.
-
integer, pointer mom_sum_output::sum_output_cs::ntrunc => NULL() The number of times the velocity has been truncated since the last call to write_energy.
-
real
max_energy¶ The maximum permitted energy per unit mass. If there is more energy than this, the model should stop [m2 s-2].
-
integer
maxtrunc¶ The number of truncations per energy save interval at which the run is stopped.
-
logical
write_stocks¶ If true, write the integrated tracer amounts to stdout when the energy files are written.
-
integer
previous_calls= 0¶ The number of times write_energy has been called.
-
integer
prev_n= 0¶ The value of n from the last call.
-
integer
fileenergy_nc¶ NetCDF id of the energy file.
-
integer
fileenergy_ascii¶ The unit number of the ascii version of the energy file.
-
type(fieldtype), dimension(num_fields+max_fields_) mom_sum_output::sum_output_cs::fields fieldtype variables for the output fields.
-
character(len=200) mom_sum_output::sum_output_cs::energyfile The name of the energy file with path.
-
- type
Pointers to various fields which may be used describe the surface state of MOM, and which will be returned to a the calling program.
Public Members
-
real, dimension(:,:), allocatable mom_variables::surface::sst The sea surface temperature [degC].
-
real, dimension(:,:), allocatable mom_variables::surface::sss The sea surface salinity [ppt ~> psu or gSalt/kg].
-
real, dimension(:,:), allocatable mom_variables::surface::sfc_density The mixed layer density [kg m-3].
-
real, dimension(:,:), allocatable mom_variables::surface::hml The mixed layer depth [m].
-
real, dimension(:,:), allocatable mom_variables::surface::u The mixed layer zonal velocity [m s-1].
-
real, dimension(:,:), allocatable mom_variables::surface::v The mixed layer meridional velocity [m s-1].
-
real, dimension(:,:), allocatable mom_variables::surface::sea_lev The sea level [m]. If a reduced surface gravity is.
-
real, dimension(:,:), allocatable mom_variables::surface::melt_potential Instantaneous amount of heat that can be used to melt sea ice [J m-2].
-
real, dimension(:,:), allocatable mom_variables::surface::ocean_mass The total mass of the ocean [kg m-2].
-
real, dimension(:,:), allocatable mom_variables::surface::ocean_heat The total heat content of the ocean in [degC kg m-2].
-
real, dimension(:,:), allocatable mom_variables::surface::ocean_salt The total salt content of the ocean in [kgSalt m-2].
-
real, dimension(:,:), allocatable mom_variables::surface::salt_deficit The salt needed to maintain the ocean column at a minimum.
-
logical
t_is_cont= .false.¶ If true, the temperature variable SST is actually the conservative temperature in [degC].
-
logical
s_is_abss= .false.¶ If true, the salinity variable SSS is actually the absolute salinity in [g/kg].
-
real, dimension(:,:), pointer mom_variables::surface::taux_shelf => NULL() The zonal stresses on the ocean under shelves [Pa].
-
real, dimension(:,:), pointer mom_variables::surface::tauy_shelf => NULL() The meridional stresses on the ocean under shelves [Pa].
-
real, dimension(:,:), pointer mom_variables::surface::frazil => NULL() The energy needed to heat the ocean column to the freezing point during the call to step_MOM [J m-2].
-
real, dimension(:,:), pointer mom_variables::surface::tempxpme => NULL() The net inflow of water into the ocean times the temperature at which this inflow occurs during the call to step_MOM [degC kg m-2]. This should be prescribed in the forcing fields, but as it often is not, this is a useful heat budget diagnostic.
-
real, dimension(:,:), pointer mom_variables::surface::internal_heat => NULL() Any internal or geothermal heat sources that are applied to the ocean integrated over the call to step_MOM [degC kg m-2].
-
type(coupler_2d_bc_type) mom_variables::surface::tr_fields A structure that may contain an array of named fields describing tracer-related quantities.
-
logical
arrays_allocated= .false.¶ A flag that indicates whether the surface type has had its memory allocated.
-
- type
A structure with diagnostic IDs of the surface and integrated variables.
Unnamed Group
-
integer
id_zos= -1¶ Diagnostic IDs for 2-d surface and bottom flux and state fields.
-
integer
id_zossq= -1¶ Diagnostic IDs for 2-d surface and bottom flux and state fields.
-
integer
id_volo= -1¶ Diagnostic IDs for 2-d surface and bottom flux and state fields.
-
integer
id_speed= -1¶ Diagnostic IDs for 2-d surface and bottom flux and state fields.
-
integer
id_ssh= -1¶ Diagnostic IDs for 2-d surface and bottom flux and state fields.
-
integer
id_ssh_ga= -1¶ Diagnostic IDs for 2-d surface and bottom flux and state fields.
-
integer
id_sst= -1¶ Diagnostic IDs for 2-d surface and bottom flux and state fields.
-
integer
id_sst_sq= -1¶ Diagnostic IDs for 2-d surface and bottom flux and state fields.
-
integer
id_sstcon= -1¶ Diagnostic IDs for 2-d surface and bottom flux and state fields.
-
integer
id_sss= -1¶ Diagnostic IDs for 2-d surface and bottom flux and state fields.
-
integer
id_sss_sq= -1¶ Diagnostic IDs for 2-d surface and bottom flux and state fields.
-
integer
id_sssabs= -1¶ Diagnostic IDs for 2-d surface and bottom flux and state fields.
-
integer
id_ssu= -1¶ Diagnostic IDs for 2-d surface and bottom flux and state fields.
-
integer
id_ssv= -1¶ Diagnostic IDs for 2-d surface and bottom flux and state fields.
-
integer
id_fraz= -1¶ Diagnostic IDs for 2-d surface and bottom flux and state fields.
-
integer
id_salt_deficit= -1¶ Diagnostic IDs for 2-d surface and bottom flux and state fields.
-
integer
id_heat_pme= -1¶ Diagnostic IDs for 2-d surface and bottom flux and state fields.
-
integer
id_intern_heat= -1¶ Diagnostic IDs for 2-d surface and bottom flux and state fields.
-
integer
- type
Structure containing pointers to the forcing fields that may be used to drive MOM. All fluxes are positive into the ocean.
Unnamed Group
-
type(user_revise_forcing_cs), pointer mom_surface_forcing::surface_forcing_cs::urf_cs => NULL() Control structures for named forcing packages.
-
type(user_surface_forcing_cs), pointer mom_surface_forcing::surface_forcing_cs::user_forcing_csp => NULL() Control structures for named forcing packages.
-
type(bfb_surface_forcing_cs), pointer mom_surface_forcing::surface_forcing_cs::bfb_forcing_csp => NULL() Control structures for named forcing packages.
-
type(dumbbell_surface_forcing_cs), pointer mom_surface_forcing::surface_forcing_cs::dumbbell_forcing_csp => NULL() Control structures for named forcing packages.
-
type(meso_surface_forcing_cs), pointer mom_surface_forcing::surface_forcing_cs::meso_forcing_csp => NULL() Control structures for named forcing packages.
-
type(neverland_surface_forcing_cs), pointer mom_surface_forcing::surface_forcing_cs::neverland_forcing_csp => NULL() Control structures for named forcing packages.
-
type(idealized_hurricane_cs), pointer mom_surface_forcing::surface_forcing_cs::idealized_hurricane_csp => NULL() Control structures for named forcing packages.
-
type(scm_cvmix_tests_cs), pointer mom_surface_forcing::surface_forcing_cs::scm_cvmix_tests_csp => NULL() Control structures for named forcing packages.
Public Members
-
logical
use_temperature¶ if true, temp & salinity used as state variables
-
logical
restorebuoy¶ if true, use restoring surface buoyancy forcing
-
logical
adiabatic¶ if true, no diapycnal mass fluxes or surface buoyancy forcing
-
logical
variable_winds¶ if true, wind stresses vary with time
-
logical
variable_buoyforce¶ if true, buoyancy forcing varies with time.
-
real
south_lat¶ southern latitude of the domain
-
real
len_lat¶ domain length in latitude
-
real
rho0¶ Boussinesq reference density [kg m-3].
-
real
g_earth¶ gravitational acceleration [L2 Z-1 T-2 ~> m s-2]
-
real
flux_const¶ piston velocity for surface restoring [m s-1]
-
real
flux_const_t¶ piston velocity for surface temperature restoring [m s-1]
-
real
flux_const_s¶ piston velocity for surface salinity restoring [m s-1]
-
real
latent_heat_fusion¶ latent heat of fusion [J kg-1]
-
real
latent_heat_vapor¶ latent heat of vaporization [J kg-1]
-
real
tau_x0¶ Constant zonal wind stress used in the WIND_CONFIG=”const” forcing.
-
real
tau_y0¶ Constant meridional wind stress used in the WIND_CONFIG=”const” forcing.
-
real
gust_const¶ constant unresolved background gustiness for ustar [Pa]
-
logical
read_gust_2d¶ if true, use 2-dimensional gustiness supplied from a file
-
real, dimension(:,:), pointer mom_surface_forcing::surface_forcing_cs::gust => NULL() spatially varying unresolved background gustiness [Pa] gust is used when read_gust_2d is true.
-
real, dimension(:,:), pointer mom_surface_forcing::surface_forcing_cs::t_restore => NULL() temperature to damp (restore) the SST to [degC]
-
real, dimension(:,:), pointer mom_surface_forcing::surface_forcing_cs::s_restore => NULL() salinity to damp (restore) the SSS [ppt]
-
real, dimension(:,:), pointer mom_surface_forcing::surface_forcing_cs::dens_restore => NULL() density to damp (restore) surface density [kg m-3]
-
integer
buoy_last_lev_read= -1¶ The last time level read from buoyancy input files.
-
real
gyres_taux_const¶ A constant wind stress [Pa].
-
real
gyres_taux_sin_amp¶ The amplitude of cosine wind stress gyres [Pa], if WIND_CONFIG==’gyres’.
-
real
gyres_taux_cos_amp¶ The amplitude of cosine wind stress gyres [Pa], if WIND_CONFIG==’gyres’.
-
real
gyres_taux_n_pis¶ The number of sine lobes in the basin if if WIND_CONFIG==’gyres’.
-
logical
answers_2018¶ If true, use the order of arithmetic and expressions that recover the answers from the end of 2018. Otherwise, use a form of the gyre wind stresses that are rotationally invariant and more likely to be the same between compilers.
-
real
t_north¶ target temperatures at north used in buoyancy_forcing_linear
-
real
t_south¶ target temperatures at south used in buoyancy_forcing_linear
-
real
s_north¶ target salinity at north used in buoyancy_forcing_linear
-
real
s_south¶ target salinity at south used in buoyancy_forcing_linear
-
logical
first_call_set_forcing= .true.¶ True until after the first call to set_forcing.
-
logical
archaic_omip_file= .true.¶ If true use the variable names and data fields from a very old version of the OMIP forcing.
-
logical
dataoverrideisinitialized= .false.¶ If true, data override has been initialized.
-
real
wind_scale¶ value by which wind-stresses are scaled, ND.
-
real
constantheatforcing¶ value used for sensible heat flux when buoy_config=”const”
-
character(len=8) mom_surface_forcing::surface_forcing_cs::wind_stagger A character indicating how the wind stress components are staggered in WIND_FILE. Valid values are A or C for now.
-
type(tracer_flow_control_cs), pointer mom_surface_forcing::surface_forcing_cs::tracer_flow_csp => NULL() A pointer to the structure that is used to orchestrate the calling of tracer packages.
-
type(mom_restart_cs), pointer mom_surface_forcing::surface_forcing_cs::restart_csp => NULL() A pointer to the restart control structure.
-
type(diag_ctrl), pointer mom_surface_forcing::surface_forcing_cs::diag structure used to regulate timing of diagnostic output
-
character(len=200) mom_surface_forcing::surface_forcing_cs::inputdir directory where NetCDF input files are.
-
character(len=200) mom_surface_forcing::surface_forcing_cs::wind_config indicator for wind forcing type (2gyre, USER, FILE..)
-
character(len=200) mom_surface_forcing::surface_forcing_cs::wind_file if wind_config is “file”, file to use
-
character(len=200) mom_surface_forcing::surface_forcing_cs::buoy_config indicator for buoyancy forcing type
-
character(len=200) mom_surface_forcing::surface_forcing_cs::longwave_file = '' The file from which the longwave heat flux is read.
-
character(len=200) mom_surface_forcing::surface_forcing_cs::shortwave_file = '' The file from which the shortwave heat flux is read.
-
character(len=200) mom_surface_forcing::surface_forcing_cs::evaporation_file = '' The file from which the evaporation is read.
-
character(len=200) mom_surface_forcing::surface_forcing_cs::sensibleheat_file = '' The file from which the sensible heat flux is read.
-
character(len=200) mom_surface_forcing::surface_forcing_cs::latentheat_file = '' The file from which the latent heat flux is read.
-
character(len=200) mom_surface_forcing::surface_forcing_cs::rain_file = '' The file from which the rainfall is read.
-
character(len=200) mom_surface_forcing::surface_forcing_cs::snow_file = '' The file from which the snowfall is read.
-
character(len=200) mom_surface_forcing::surface_forcing_cs::runoff_file = '' The file from which the runoff is read.
-
character(len=200) mom_surface_forcing::surface_forcing_cs::longwaveup_file = '' The file from which the upward longwave heat flux is read.
-
character(len=200) mom_surface_forcing::surface_forcing_cs::shortwaveup_file = '' The file from which the upward shorwave heat flux is read.
-
character(len=200) mom_surface_forcing::surface_forcing_cs::sstrestore_file = '' The file from which to read the sea surface temperature to restore toward.
-
character(len=200) mom_surface_forcing::surface_forcing_cs::salinityrestore_file = '' The file from which to read the sea surface salinity to restore toward.
-
character(len=80) mom_surface_forcing::surface_forcing_cs::stress_x_var = '' X-windstress variable name in the input file.
-
character(len=80) mom_surface_forcing::surface_forcing_cs::stress_y_var = '' Y-windstress variable name in the input file.
-
character(len=80) mom_surface_forcing::surface_forcing_cs::ustar_var = '' ustar variable name in the input file
-
character(len=80) mom_surface_forcing::surface_forcing_cs::lw_var = '' lonngwave heat flux variable name in the input file
-
character(len=80) mom_surface_forcing::surface_forcing_cs::sw_var = '' shortwave heat flux variable name in the input file
-
character(len=80) mom_surface_forcing::surface_forcing_cs::latent_var = '' latent heat flux variable name in the input file
-
character(len=80) mom_surface_forcing::surface_forcing_cs::sens_var = '' sensible heat flux variable name in the input file
-
character(len=80) mom_surface_forcing::surface_forcing_cs::evap_var = '' evaporation variable name in the input file
-
character(len=80) mom_surface_forcing::surface_forcing_cs::rain_var = '' rainfall variable name in the input file
-
character(len=80) mom_surface_forcing::surface_forcing_cs::snow_var = '' snowfall variable name in the input file
-
character(len=80) mom_surface_forcing::surface_forcing_cs::lrunoff_var = '' liquid runoff variable name in the input file
-
character(len=80) mom_surface_forcing::surface_forcing_cs::frunoff_var = '' frozen runoff variable name in the input file
-
character(len=80) mom_surface_forcing::surface_forcing_cs::sst_restore_var = '' target sea surface temeperature variable name in the input file
-
character(len=80) mom_surface_forcing::surface_forcing_cs::sss_restore_var = '' target sea surface salinity variable name in the input file
-
integer
wind_nlev= -1¶ The number of time levels in the file of wind stress.
-
integer
sw_nlev= -1¶ The number of time levels in the file of shortwave heat flux.
-
integer
lw_nlev= -1¶ The number of time levels in the file of longwave heat flux.
-
integer
latent_nlev= -1¶ The number of time levels in the file of latent heat flux.
-
integer
sens_nlev= -1¶ The number of time levels in the file of sensible heat flux.
-
integer
evap_nlev= -1¶ The number of time levels in the file of evaporation.
-
integer
precip_nlev= -1¶ The number of time levels in the file of precipitation.
-
integer
runoff_nlev= -1¶ The number of time levels in the file of runoff.
-
integer
sst_nlev= -1¶ The number of time levels in the file of target SST.
-
integer
sss_nlev= -1¶ The number of time levels in the file of target SSS.
-
integer
wind_last_lev= -1¶ The last time level read of wind stress.
-
integer
sw_last_lev= -1¶ The last time level read of shortwave heat flux.
-
integer
lw_last_lev= -1¶ The last time level read of longwave heat flux.
-
integer
latent_last_lev= -1¶ The last time level read of latent heat flux.
-
integer
sens_last_lev= -1¶ The last time level read of sensible heat flux.
-
integer
evap_last_lev= -1¶ The last time level read of evaporation.
-
integer
precip_last_lev= -1¶ The last time level read of precipitation.
-
integer
runoff_last_lev= -1¶ The last time level read of runoff.
-
integer
sst_last_lev= -1¶ The last time level read of target SST.
-
integer
sss_last_lev= -1¶ The last time level read of target SSS.
-
type(forcing_diags), public mom_surface_forcing::surface_forcing_cs::handles A structure with diagnostics handles.
-
- type
Pointers to an assortment of thermodynamic fields that may be available, including potential temperature, salinity, heat capacity, and the equation of state control structure.
Public Members
-
real, dimension(:,:,:), pointer mom_variables::thermo_var_ptrs::t => NULL() Potential temperature [degC].
-
real, dimension(:,:,:), pointer mom_variables::thermo_var_ptrs::s => NULL() Salnity [PSU] or [gSalt/kg], generically [ppt].
-
type(eos_type), pointer mom_variables::thermo_var_ptrs::eqn_of_state => NULL() Type that indicates the equation of state to use.
-
real
p_ref¶ The coordinate-density reference pressure [Pa]. This is the pressure used to calculate Rml from T and S when eqn_of_state is associated.
-
real
c_p¶ The heat capacity of seawater [J degC-1 kg-1]. When conservative temperature is used, this is constant and exactly 3991.86795711963 J degC-1 kg-1.
-
logical
t_is_cont= .false.¶ If true, the temperature variable tvT is actually the conservative temperature [degC].
-
logical
s_is_abss= .false.¶ If true, the salinity variable tvS is actually the absolute salinity in units of [gSalt/kg].
-
real
min_salinity= 0.01¶ The minimum value of salinity when BOUND_SALINITY=True [ppt]. The default is 0.01 for backward compatibility but should be 0.
-
real, dimension(:,:), pointer mom_variables::thermo_var_ptrs::frazil => NULL() The energy needed to heat the ocean column to the freezing point since calculate_surface_state was2 last called [J m-2].
-
real, dimension(:,:), pointer mom_variables::thermo_var_ptrs::salt_deficit => NULL() The salt needed to maintain the ocean column at a minimum salinity of MIN_SALINITY since the last time that calculate_surface_state was called, [gSalt m-2].
-
real, dimension(:,:), pointer mom_variables::thermo_var_ptrs::tempxpme => NULL() The net inflow of water into the ocean times the temperature at which this inflow occurs since the last call to calculate_surface_state [degC kg m-2]. This should be prescribed in the forcing fields, but as it often is not, this is a useful heat budget diagnostic.
-
real, dimension(:,:), pointer mom_variables::thermo_var_ptrs::internal_heat => NULL() Any internal or geothermal heat sources that have been applied to the ocean since the last call to calculate_surface_state [degC kg m-2].
-
- type
Control structure for thickness diffusion.
Unnamed Group
-
integer
id_uhgm= -1¶ Diagnostic identifier.
-
integer
id_vhgm= -1¶ Diagnostic identifier.
-
integer
id_gmwork= -1¶ Diagnostic identifier.
-
integer
id_kh_u= -1¶ Diagnostic identifier.
-
integer
id_kh_v= -1¶ Diagnostic identifier.
-
integer
id_kh_t= -1¶ Diagnostic identifier.
-
integer
id_kh_u1= -1¶ Diagnostic identifier.
-
integer
id_kh_v1= -1¶ Diagnostic identifier.
-
integer
id_kh_t1= -1¶ Diagnostic identifier.
-
integer
id_slope_x= -1¶ Diagnostic identifier.
-
integer
id_slope_y= -1¶ Diagnostic identifier.
-
integer
id_sfn_unlim_x= -1¶ Diagnostic identifier.
-
integer
id_sfn_unlim_y= -1¶ Diagnostic identifier.
-
integer
id_sfn_x= -1¶ Diagnostic identifier.
-
integer
id_sfn_y= -1¶ Diagnostic identifier.
Public Members
-
real
khth¶ Background interface depth diffusivity [m2 s-1].
-
real
khth_slope_cff¶ Slope dependence coefficient of Khth [m2 s-1].
-
real
max_khth_cfl¶ Maximum value of the diffusive CFL for thickness diffusion.
-
real
khth_min¶ Minimum value of Khth [m2 s-1].
-
real
khth_max¶ Maximum value of Khth [m2 s-1], or 0 for no max.
-
real
slope_max¶ Slopes steeper than slope_max are limited in some way [nondim].
-
real
kappa_smooth¶ Vertical diffusivity used to interpolate more sensible values of T & S into thin layers [Z2 s-1 ~> m2 s-1].
-
logical
thickness_diffuse¶ If true, interfaces heights are diffused.
-
logical
use_fgnv_streamfn¶ If true, use the streamfunction formulation of Ferrari et al., 2010, which effectively emphasizes graver vertical modes by smoothing in the vertical.
-
real
fgnv_scale¶ A coefficient scaling the vertical smoothing term in the Ferrari et al., 2010, streamfunction formulation [nondim].
-
real
fgnv_c_min¶ A minimum wave speed used in the Ferrari et al., 2010, streamfunction formulation [m s-1].
-
real
n2_floor¶ A floor for Brunt-Vasaila frequency in the Ferrari et al., 2010, streamfunction formulation [s-2].
-
logical
detangle_interfaces¶ If true, add 3-d structured interface height diffusivities to horizontally smooth jagged layers.
-
real
detangle_time¶ If detangle_interfaces is true, this is the timescale over which maximally jagged grid-scale thickness variations are suppressed [s]. This must be longer than DT, or 0 (the default) to use DT.
-
integer
nkml¶ number of layers within mixed layer
-
logical
debug¶ write verbose checksums for debugging purposes
-
logical
use_gme_thickness_diffuse¶ If true, passes GM coefficients to MOM_hor_visc for use with GME closure.
-
logical
meke_geometric¶ If true, uses the GM coefficient formulation from the GEOMETRIC framework (Marshall et al., 2012)
-
real
meke_geometric_alpha¶ The nondimensional coefficient governing the efficiency of the GEOMETRIC thickness difussion [nondim].
-
real
meke_geometric_epsilon¶ Minimum Eady growth rate for the GEOMETRIC thickness diffusivity [s-1].
-
logical
use_kh_in_meke¶ If true, uses the thickness diffusivity calculated here to diffuse MEKE.
-
logical
gm_src_alt¶ If true, use the GM energy conversion form S^2*N^2*kappa rather than the streamfunction for the GM source term.
-
type(diag_ctrl), pointer mom_thickness_diffuse::thickness_diffuse_cs::diag => NULL() structure used to regulate timing of diagnostics
-
real, dimension(:,:), pointer mom_thickness_diffuse::thickness_diffuse_cs::gmwork => NULL() Work by thickness diffusivity [W m-2].
-
real, dimension(:,:,:), pointer mom_thickness_diffuse::thickness_diffuse_cs::diagslopex => NULL() Diagnostic: zonal neutral slope [nondim].
-
real, dimension(:,:,:), pointer mom_thickness_diffuse::thickness_diffuse_cs::diagslopey => NULL() Diagnostic: zonal neutral slope [nondim].
-
real, dimension(:,:,:), pointer mom_thickness_diffuse::thickness_diffuse_cs::kh_u_gme => NULL() interface height diffusivities in u-columns (m2 s-1)
-
real, dimension(:,:,:), pointer mom_thickness_diffuse::thickness_diffuse_cs::kh_v_gme => NULL() interface height diffusivities in v-columns (m2 s-1)
-
integer
- type
Control structure for tidal bay open boundaries.
Public Members
-
real
tide_flow= 3.0e6¶ Maximum tidal flux.
-
real
- type
The control structure for the MOM_tidal_forcing module.
Public Members
-
logical
use_sal_scalar¶ If true, use the scalar approximation when calculating self-attraction and loading.
-
logical
tidal_sal_from_file¶ If true, Read the tidal self-attraction and loading from input files, specified by TIDAL_INPUT_FILE.
-
logical
use_prev_tides¶ If true, use the SAL from the previous iteration of the tides to facilitate convergence.
-
real
sal_scalar¶ The constant of proportionality between sea surface height (really it should be bottom pressure) anomalies and bottom geopotential anomalies.
-
integer
nc¶ The number of tidal constituents in use.
-
real, dimension(max_constituents) mom_tidal_forcing::tidal_forcing_cs::freq The frequency of a tidal constituent [s-1].
-
real, dimension(max_constituents) mom_tidal_forcing::tidal_forcing_cs::phase0 The phase of a tidal constituent at time 0, in radians.
-
real, dimension(max_constituents) mom_tidal_forcing::tidal_forcing_cs::amp The amplitude of a tidal constituent at time 0 [m].
-
real, dimension(max_constituents) mom_tidal_forcing::tidal_forcing_cs::love_no The Love number of a tidal constituent at time 0 [nondim].
-
integer, dimension(max_constituents) mom_tidal_forcing::tidal_forcing_cs::struct An encoded spatial structure for each constituent.
-
character (len=16), dimension(max_constituents) mom_tidal_forcing::tidal_forcing_cs::const_name The name of each constituent.
-
real, dimension(:,:,:), pointer mom_tidal_forcing::tidal_forcing_cs::sin_struct => NULL() The sine and cosine based structures that can.
-
real, dimension(:,:,:), pointer mom_tidal_forcing::tidal_forcing_cs::cos_struct => NULL() be associated with the astronomical forcing.
-
real, dimension(:,:,:), pointer mom_tidal_forcing::tidal_forcing_cs::cosphasesal => NULL() The cosine and sine of the phase of the.
-
real, dimension(:,:,:), pointer mom_tidal_forcing::tidal_forcing_cs::sinphasesal => NULL() self-attraction and loading amphidromes.
-
real, dimension(:,:,:), pointer mom_tidal_forcing::tidal_forcing_cs::ampsal => NULL() The amplitude of the SAL [m].
-
real, dimension(:,:,:), pointer mom_tidal_forcing::tidal_forcing_cs::cosphase_prev => NULL() The cosine and sine of the phase of the.
-
real, dimension(:,:,:), pointer mom_tidal_forcing::tidal_forcing_cs::sinphase_prev => NULL() amphidromes in the previous tidal solutions.
-
real, dimension(:,:,:), pointer mom_tidal_forcing::tidal_forcing_cs::amp_prev => NULL() The amplitude of the previous tidal solution [m].
-
logical
- type
Control structure with parameters for the tidal mixing module.
Unnamed Group
-
integer
id_tke_itidal= -1¶ Diagnostic identifiers.
-
integer
id_tke_leewave= -1¶ Diagnostic identifiers.
-
integer
id_kd_itidal= -1¶ Diagnostic identifiers.
-
integer
id_kd_niku= -1¶ Diagnostic identifiers.
-
integer
id_kd_lowmode= -1¶ Diagnostic identifiers.
-
integer
id_kd_itidal_work= -1¶ Diagnostic identifiers.
-
integer
id_kd_niku_work= -1¶ Diagnostic identifiers.
-
integer
id_kd_lowmode_work= -1¶ Diagnostic identifiers.
-
integer
id_nb= -1¶ Diagnostic identifiers.
-
integer
id_n2_bot= -1¶ Diagnostic identifiers.
-
integer
id_n2_meanz= -1¶ Diagnostic identifiers.
-
integer
id_fl_itidal= -1¶ Diagnostic identifiers.
-
integer
id_fl_lowmode= -1¶ Diagnostic identifiers.
-
integer
id_polzin_decay_scale= -1¶ Diagnostic identifiers.
-
integer
id_polzin_decay_scale_scaled= -1¶ Diagnostic identifiers.
-
integer
id_n2_int= -1¶ Diagnostic identifiers.
-
integer
id_simmons_coeff= -1¶ Diagnostic identifiers.
-
integer
id_schmittner_coeff= -1¶ Diagnostic identifiers.
-
integer
id_tidal_qe_md= -1¶ Diagnostic identifiers.
-
integer
id_vert_dep= -1¶ Diagnostic identifiers.
Public Members
-
logical
debug= .true.¶ If true, do more extensive debugging checks. This is hard-coded.
-
logical
int_tide_dissipation= .false.¶ Internal tide conversion (from barotropic) with the schemes of St Laurent et al (2002) & Simmons et al (2004)
-
integer
int_tide_profile¶ A coded integer indicating the vertical profile for dissipation of the internal waves. Schemes that are currently encoded are St Laurent et al (2002) and Polzin (2009).
-
logical
lee_wave_dissipation= .false.¶ Enable lee-wave driven mixing, following Nikurashin (2010), with a vertical energy deposition profile specified by Lee_wave_profile to be St Laurent et al (2002) or Simmons et al (2004) scheme.
-
integer
lee_wave_profile¶ A coded integer indicating the vertical profile for dissipation of the lee waves. Schemes that are currently encoded are St Laurent et al (2002) and Polzin (2009).
-
real
int_tide_decay_scale¶ decay scale for internal wave TKE [Z ~> m].
-
real
mu_itides¶ efficiency for conversion of dissipation to potential energy [nondim]
-
real
gamma_itides¶ fraction of local dissipation [nondim]
-
real
gamma_lee¶ fraction of local dissipation for lee waves (Nikurashin’s energy input) [nondim]
-
real
decay_scale_factor_lee¶ Scaling factor for the decay scale of lee wave energy dissipation [nondim].
-
real
min_zbot_itides¶ minimum depth for internal tide conversion [Z ~> m].
-
logical
lowmode_itidal_dissipation= .false.¶ If true, consider mixing due to breaking low modes that have been remotely generated using an internal tidal dissipation scheme to specify the vertical profile of the energy input to drive diapycnal mixing, along the lines of St. Laurent et al. (2002) and Simmons et al. (2004).
-
real
nu_polzin¶ The non-dimensional constant used in Polzin form of the vertical scale of decay of tidal dissipation.
-
real
nbotref_polzin¶ Reference value for the buoyancy frequency at the ocean bottom used in Polzin formulation of the vertical scale of decay of tidal dissipation [T-1 ~> s-1].
-
real
polzin_decay_scale_factor¶ Scaling factor for the decay length scale of the tidal dissipation profile in Polzin [nondim].
-
real
polzin_decay_scale_max_factor¶ The decay length scale of tidal dissipation profile in Polzin formulation should not exceed Polzin_decay_scale_max_factor * depth of the ocean [nondim].
-
real
polzin_min_decay_scale¶ minimum decay scale of the tidal dissipation profile in Polzin formulation [Z ~> m].
-
real
tke_itide_max¶ maximum internal tide conversion [kg Z3 m-3 T-3 ~> W m-2] available to mix above the BBL
-
real
utide¶ constant tidal amplitude [Z T-1 ~> m s-1] if READ_TIDEAMP is false.
-
real
kappa_itides¶ topographic wavenumber and non-dimensional scaling [Z-1 ~> m-1].
-
real
kappa_h2_factor¶ factor for the product of wavenumber * rms sgs height
-
character(len=200) mom_tidal_mixing::tidal_mixing_cs::inputdir The directory in which to find input files.
-
logical
use_cvmix_tidal= .false.¶ true if CVMix is to be used for determining diffusivity due to tidal mixing
-
real
min_thickness¶ Minimum thickness allowed [m].
-
integer
cvmix_tidal_scheme= -1¶ 1 for Simmons, 2 for Schmittner
-
type(cvmix_tidal_params_type) mom_tidal_mixing::tidal_mixing_cs::cvmix_tidal_params A CVMix-specific type with parameters for tidal mixing.
-
type(cvmix_global_params_type) mom_tidal_mixing::tidal_mixing_cs::cvmix_glb_params CVMix-specific for Prandtl number only.
-
real
tidal_max_coef¶ CVMix-specific maximum allowable tidal diffusivity. [m^2/s].
-
real
tidal_diss_lim_tc¶ CVMix-specific dissipation limit depth for tidal-energy-constituent data [Z ~> m].
-
type(remapping_cs) mom_tidal_mixing::tidal_mixing_cs::remap_cs The control structure for remapping.
-
real, dimension(:,:), pointer mom_tidal_mixing::tidal_mixing_cs::tke_niku => NULL() Lee wave driven Turbulent Kinetic Energy input [kg Z3 m-3 T-3 ~> W m-2].
-
real, dimension(:,:), pointer mom_tidal_mixing::tidal_mixing_cs::tke_itidal => NULL() The internal Turbulent Kinetic Energy input divided by the bottom stratfication [kg Z3 m-3 T-2 ~> J m-2].
-
real, dimension(:,:), pointer mom_tidal_mixing::tidal_mixing_cs::nb => NULL() The near bottom buoyancy frequency [T-1 ~> s-1].
-
real, dimension(:,:), pointer mom_tidal_mixing::tidal_mixing_cs::mask_itidal => NULL() A mask of where internal tide energy is input.
-
real, dimension(:,:), pointer mom_tidal_mixing::tidal_mixing_cs::h2 => NULL() Squared bottom depth variance [m2].
-
real, dimension(:,:), pointer mom_tidal_mixing::tidal_mixing_cs::tideamp => NULL() RMS tidal amplitude [m s-1].
-
real, dimension(:), allocatable mom_tidal_mixing::tidal_mixing_cs::h_src tidal constituent input layer thickness [m]
-
real, dimension(:,:), allocatable mom_tidal_mixing::tidal_mixing_cs::tidal_qe_2d Tidal energy input times the local dissipation fraction, q*E(x,y), with the CVMix implementation of Jayne et al tidal mixing [W m-2]. TODO: make this E(x,y) only.
-
real, dimension(:,:,:), allocatable mom_tidal_mixing::tidal_mixing_cs::tidal_qe_3d_in q*E(x,y,z) with the Schmittner parameterization [W m-3?]
-
logical
answers_2018¶ If true, use the order of arithmetic and expressions that recover the answers from the end of 2018. Otherwise, use updated and more robust forms of the same expressions.
-
type(diag_ctrl), pointer mom_tidal_mixing::tidal_mixing_cs::diag => NULL() structure to regulate diagnostic output timing
-
type(tidal_mixing_diags), pointer mom_tidal_mixing::tidal_mixing_cs::dd => NULL() A pointer to a structure of diagnostic arrays.
-
integer
- type
Containers for tidal mixing diagnostics.
Public Members
-
real, dimension(:,:,:), pointer mom_tidal_mixing::tidal_mixing_diags::kd_itidal => NULL() internal tide diffusivity at interfaces [Z2 T-1 ~> m2 s-1].
-
real, dimension(:,:,:), pointer mom_tidal_mixing::tidal_mixing_diags::fl_itidal => NULL() vertical flux of tidal turbulent dissipation [Z3 T-3 ~> m3 s-3]
-
real, dimension(:,:,:), pointer mom_tidal_mixing::tidal_mixing_diags::kd_niku => NULL() lee-wave diffusivity at interfaces [Z2 T-1 ~> m2 s-1].
-
real, dimension(:,:,:), pointer mom_tidal_mixing::tidal_mixing_diags::kd_niku_work => NULL() layer integrated work by lee-wave driven mixing [kg Z3 m-3 T-3 ~> W m-2]
-
real, dimension(:,:,:), pointer mom_tidal_mixing::tidal_mixing_diags::kd_itidal_work => NULL() layer integrated work by int tide driven mixing [kg Z3 m-3 T-3 ~> W m-2]
-
real, dimension(:,:,:), pointer mom_tidal_mixing::tidal_mixing_diags::kd_lowmode_work => NULL() layer integrated work by low mode driven mixing [kg Z3 m-3 T-3 ~> W m-2]
-
real, dimension(:,:,:), pointer mom_tidal_mixing::tidal_mixing_diags::n2_int => NULL() Bouyancy frequency squared at interfaces [s-2].
-
real, dimension(:,:,:), pointer mom_tidal_mixing::tidal_mixing_diags::vert_dep_3d => NULL() The 3-d mixing energy deposition [W m-3].
-
real, dimension(:,:,:), pointer mom_tidal_mixing::tidal_mixing_diags::schmittner_coeff_3d => NULL() The coefficient in the Schmittner et al mixing scheme, in UNITS?
-
real, dimension(:,:,:), pointer mom_tidal_mixing::tidal_mixing_diags::tidal_qe_md => NULL() Input tidal energy dissipated locally, interpolated to model vertical coordinate [W m-3?].
-
real, dimension(:,:,:), pointer mom_tidal_mixing::tidal_mixing_diags::kd_lowmode => NULL() internal tide diffusivity at interfaces due to propagating low modes [Z2 T-1 ~> m2 s-1].
-
real, dimension(:,:,:), pointer mom_tidal_mixing::tidal_mixing_diags::fl_lowmode => NULL() vertical flux of tidal turbulent dissipation due to propagating low modes [Z3 T-3 ~> m3 s-3]
-
real, dimension(:,:), pointer mom_tidal_mixing::tidal_mixing_diags::tke_itidal_used => NULL() internal tide TKE input at ocean bottom [kg Z3 m-3 T-3 ~> W m-2]
-
real, dimension(:,:), pointer mom_tidal_mixing::tidal_mixing_diags::n2_bot => NULL() bottom squared buoyancy frequency [T-2 ~> s-2]
-
real, dimension(:,:), pointer mom_tidal_mixing::tidal_mixing_diags::n2_meanz => NULL() vertically averaged buoyancy frequency [T-2 ~> s-2]
-
real, dimension(:,:), pointer mom_tidal_mixing::tidal_mixing_diags::polzin_decay_scale_scaled => NULL() vertical scale of decay for tidal dissipation
-
real, dimension(:,:), pointer mom_tidal_mixing::tidal_mixing_diags::polzin_decay_scale => NULL() vertical decay scale for tidal diss with Polzin [m]
-
real, dimension(:,:), pointer mom_tidal_mixing::tidal_mixing_diags::simmons_coeff_2d => NULL() The Simmons et al mixing coefficient.
-
- type
Control structure for this module.
Public Members
-
real
dt¶ The baroclinic dynamics time step [s].
-
type(diag_ctrl), pointer mom_tracer_advect::tracer_advect_cs::diag A structure that is used to regulate the timing of diagnostic output.
-
logical
debug¶ If true, write verbose checksums for debugging purposes.
-
logical
useppm¶ If true, use PPM instead of PLM.
-
logical
usehuynh¶ If true, use the Huynh scheme for PPM interface values.
-
type(group_pass_type) mom_tracer_advect::tracer_advect_cs::pass_uhr_vhr_t_hprev A structred used for group passes.
-
real
- type
The control structure for orchestrating the calling of tracer packages.
Unnamed Group
-
type(user_tracer_example_cs), pointer mom_tracer_flow_control::tracer_flow_control_cs::user_tracer_example_csp => NULL() Pointers to the control strucures for the tracer packages.
-
type(dome_tracer_cs), pointer mom_tracer_flow_control::tracer_flow_control_cs::dome_tracer_csp => NULL() Pointers to the control strucures for the tracer packages.
-
type(isomip_tracer_cs), pointer mom_tracer_flow_control::tracer_flow_control_cs::isomip_tracer_csp => NULL() Pointers to the control strucures for the tracer packages.
-
type(rgc_tracer_cs), pointer mom_tracer_flow_control::tracer_flow_control_cs::rgc_tracer_csp => NULL() Pointers to the control strucures for the tracer packages.
-
type(ideal_age_tracer_cs), pointer mom_tracer_flow_control::tracer_flow_control_cs::ideal_age_tracer_csp => NULL() Pointers to the control strucures for the tracer packages.
-
type(dye_tracer_cs), pointer mom_tracer_flow_control::tracer_flow_control_cs::dye_tracer_csp => NULL() Pointers to the control strucures for the tracer packages.
-
type(oil_tracer_cs), pointer mom_tracer_flow_control::tracer_flow_control_cs::oil_tracer_csp => NULL() Pointers to the control strucures for the tracer packages.
-
type(advection_test_tracer_cs), pointer mom_tracer_flow_control::tracer_flow_control_cs::advection_test_tracer_csp => NULL() Pointers to the control strucures for the tracer packages.
-
type(ocmip2_cfc_cs), pointer mom_tracer_flow_control::tracer_flow_control_cs::ocmip2_cfc_csp => NULL() Pointers to the control strucures for the tracer packages.
-
type(pseudo_salt_tracer_cs), pointer mom_tracer_flow_control::tracer_flow_control_cs::pseudo_salt_tracer_csp => NULL() Pointers to the control strucures for the tracer packages.
-
type(boundary_impulse_tracer_cs), pointer mom_tracer_flow_control::tracer_flow_control_cs::boundary_impulse_tracer_csp => NULL() Pointers to the control strucures for the tracer packages.
-
type(dyed_obc_tracer_cs), pointer mom_tracer_flow_control::tracer_flow_control_cs::dyed_obc_tracer_csp => NULL() Pointers to the control strucures for the tracer packages.
Public Members
-
logical
use_user_tracer_example= .false.¶ If true, use the USER_tracer_example package.
-
logical
use_dome_tracer= .false.¶ If true, use the DOME_tracer package.
-
logical
use_isomip_tracer= .false.¶ If true, use the ISOMPE_tracer package.
-
logical
use_rgc_tracer= .false.¶ If true, use the RGC_tracer package.
-
logical
use_ideal_age= .false.¶ If true, use the ideal age tracer package.
-
logical
use_regional_dyes= .false.¶ If true, use the regional dyes tracer package.
-
logical
use_oil= .false.¶ If true, use the oil tracer package.
-
logical
use_advection_test_tracer= .false.¶ If true, use the advection_test_tracer package.
-
logical
use_ocmip2_cfc= .false.¶ If true, use the OCMIP2_CFC tracer package.
-
logical
use_mom_generic_tracer= .false.¶ If true, use the MOM_generic_tracer packages.
-
logical
use_pseudo_salt_tracer= .false.¶ If true, use the psuedo_salt tracer package.
-
logical
use_boundary_impulse_tracer= .false.¶ If true, use the boundary impulse tracer package.
-
logical
use_dyed_obc_tracer= .false.¶ If true, use the dyed OBC tracer package.
-
- type
The ocntrol structure for along-layer and epineutral tracer diffusion.
Unnamed Group
-
integer
id_khtr_u= -1¶ Diagnostic IDs.
-
integer
id_khtr_v= -1¶ Diagnostic IDs.
-
integer
id_khtr_h= -1¶ Diagnostic IDs.
-
integer
id_cfl= -1¶ Diagnostic IDs.
-
integer
id_khdt_x= -1¶ Diagnostic IDs.
-
integer
id_khdt_y= -1¶ Diagnostic IDs.
-
type(group_pass_type) mom_tracer_hor_diff::tracer_hor_diff_cs::pass_t For group halo pass, used in both tracer_hordiff and tracer_epipycnal_ML_diff.
Public Members
-
real
dt¶ The baroclinic dynamics time step [s].
-
real
khtr¶ The along-isopycnal tracer diffusivity [m2 s-1].
-
real
khtr_slope_cff¶ The non-dimensional coefficient in KhTr formula.
-
real
khtr_min¶ Minimum along-isopycnal tracer diffusivity [m2 s-1].
-
real
khtr_max¶ Maximum along-isopycnal tracer diffusivity [m2 s-1].
-
real
khtr_passivity_coeff¶ Passivity coefficient that scales Rd/dx (default = 0) where passivity is the ratio between along-isopycnal tracer mixing and thickness mixing [nondim].
-
real
khtr_passivity_min¶ Passivity minimum (default = 1/2) [nondim].
-
real
ml_khtr_scale¶ With Diffuse_ML_interior, the ratio of the truly horizontal diffusivity in the mixed layer to the epipycnal diffusivity [nondim].
-
real
max_diff_cfl¶ If positive, locally limit the along-isopycnal tracer diffusivity to keep the diffusive CFL locally at or below this value [nondim].
-
logical
diffuse_ml_interior¶ If true, diffuse along isopycnals between the mixed layer and the interior.
-
logical
check_diffusive_cfl¶ If true, automatically iterate the diffusion to ensure that the diffusive equivalent of the CFL limit is not violated.
-
logical
use_neutral_diffusion¶ If true, use the neutral_diffusion module from within tracer_hor_diff.
-
type(neutral_diffusion_cs), pointer mom_tracer_hor_diff::tracer_hor_diff_cs::neutral_diffusion_csp => NULL() Control structure for neutral diffusion.
-
type(diag_ctrl), pointer mom_tracer_hor_diff::tracer_hor_diff_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
logical
debug¶ If true, write verbose checksums for debugging purposes.
-
logical
show_call_tree¶ Display the call tree while running. Set by VERBOSITY level.
-
logical
first_call= .true.¶ This is true until after the first call.
-
integer
- type
Type to carry basic tracer information.
Unnamed Group
-
integer
ntr= 0¶ number of registered tracers
-
type(tracer_type), dimension( 50 ) mom_tracer_registry::tracer_registry_type::tr array of registered tracers
-
logical
locked= .false.¶ New tracers may be registered if locked=.false. When locked=.true., no more tracers can be registered, at which point common diagnostics can be set up for the registered tracers.
-
integer
- type
The tracer type.
Unnamed Group
-
integer
id_tr= -1¶ Diagnostic IDs.
-
integer
id_adx= -1¶ Diagnostic IDs.
-
integer
id_ady= -1¶ Diagnostic IDs.
-
integer
id_dfx= -1¶ Diagnostic IDs.
-
integer
id_dfy= -1¶ Diagnostic IDs.
-
integer
id_adx_2d= -1¶ Diagnostic IDs.
-
integer
id_ady_2d= -1¶ Diagnostic IDs.
-
integer
id_dfx_2d= -1¶ Diagnostic IDs.
-
integer
id_dfy_2d= -1¶ Diagnostic IDs.
-
integer
id_adv_xy= -1¶ Diagnostic IDs.
-
integer
id_adv_xy_2d= -1¶ Diagnostic IDs.
-
integer
id_dfxy_cont= -1¶ Diagnostic IDs.
-
integer
id_dfxy_cont_2d= -1¶ Diagnostic IDs.
-
integer
id_dfxy_conc= -1¶ Diagnostic IDs.
-
integer
id_remap_conc= -1¶ Diagnostic IDs.
-
integer
id_remap_cont= -1¶ Diagnostic IDs.
-
integer
id_remap_cont_2d= -1¶ Diagnostic IDs.
-
integer
id_tendency= -1¶ Diagnostic IDs.
-
integer
id_trxh_tendency= -1¶ Diagnostic IDs.
-
integer
id_trxh_tendency_2d= -1¶ Diagnostic IDs.
-
integer
id_tr_vardec= -1¶ Diagnostic IDs.
Public Members
-
real, dimension(:,:,:), pointer mom_tracer_registry::tracer_type::t => NULL() tracer concentration array [conc]
-
real, dimension(:,:,:), pointer mom_tracer_registry::tracer_type::ad_x => NULL() diagnostic array for x-advective tracer flux [conc H m2 s-1 ~> conc m3 s-1 or conc kg s-1]
-
real, dimension(:,:,:), pointer mom_tracer_registry::tracer_type::ad_y => NULL() diagnostic array for y-advective tracer flux [conc H m2 s-1 ~> conc m3 s-1 or conc kg s-1]
-
real, dimension(:,:), pointer mom_tracer_registry::tracer_type::ad2d_x => NULL() diagnostic vertical sum x-advective tracer flux [conc H m2 s-1 ~> conc m3 s-1 or conc kg s-1]
-
real, dimension(:,:), pointer mom_tracer_registry::tracer_type::ad2d_y => NULL() diagnostic vertical sum y-advective tracer flux [conc H m2 s-1 ~> conc m3 s-1 or conc kg s-1]
-
real, dimension(:,:,:), pointer mom_tracer_registry::tracer_type::df_x => NULL() diagnostic array for x-diffusive tracer flux [conc H m2 s-1 ~> conc m3 s-1 or conc kg s-1]
-
real, dimension(:,:,:), pointer mom_tracer_registry::tracer_type::df_y => NULL() diagnostic array for y-diffusive tracer flux [conc H m2 s-1 ~> conc m3 s-1 or conc kg s-1]
-
real, dimension(:,:), pointer mom_tracer_registry::tracer_type::df2d_x => NULL() diagnostic vertical sum x-diffusive flux [conc H m2 s-1 ~> conc m3 s-1 or conc kg s-1]
-
real, dimension(:,:), pointer mom_tracer_registry::tracer_type::df2d_y => NULL() diagnostic vertical sum y-diffusive flux [conc H m2 s-1 ~> conc m3 s-1 or conc kg s-1]
-
real, dimension(:,:), pointer mom_tracer_registry::tracer_type::df2d_conc_x => NULL() diagnostic vertical sum x-diffusive content flux [conc H m2 s-1 ~> conc m3 s-1 or conc kg s-1]
-
real, dimension(:,:), pointer mom_tracer_registry::tracer_type::df2d_conc_y => NULL() diagnostic vertical sum y-diffusive content flux [conc H m2 s-1 ~> conc m3 s-1 or conc kg s-1]
-
real, dimension(:,:,:), pointer mom_tracer_registry::tracer_type::advection_xy => NULL() convergence of lateral advective tracer fluxes [conc H s-1 ~> conc m s-1 or conc kg m-2 s-1]
-
real, dimension(:,:,:), pointer mom_tracer_registry::tracer_type::diff_cont_xy => NULL() convergence of lateral diffusive tracer fluxes [conc H s-1 ~> conc m s-1 or conc kg m-2 s-1]
-
real, dimension(:,:,:), pointer mom_tracer_registry::tracer_type::diff_conc_xy => NULL() convergence of lateral diffusive tracer fluxes expressed as a change in concentration [conc s-1]
-
real, dimension(:,:,:), pointer mom_tracer_registry::tracer_type::t_prev => NULL() tracer concentration array at a previous timestep used for diagnostics [conc]
-
real, dimension(:,:,:), pointer mom_tracer_registry::tracer_type::trxh_prev => NULL() layer integrated tracer concentration array at a previous timestep used for diagnostics
-
character(len=32) mom_tracer_registry::tracer_type::name tracer name used for diagnostics and error messages
-
character(len=64) mom_tracer_registry::tracer_type::units Physical dimensions of the tracer concentration.
-
character(len=240) mom_tracer_registry::tracer_type::longname Long name of the variable.
-
logical
registry_diags= .false.¶ If true, use the registry to set up the diagnostics associated with this tracer.
-
character(len=64) mom_tracer_registry::tracer_type::cmor_name CMOR name of this tracer.
-
character(len=64) mom_tracer_registry::tracer_type::cmor_units CMOR physical dimensions of the tracer.
-
character(len=240) mom_tracer_registry::tracer_type::cmor_longname CMOR long name of the tracer.
-
character(len=32) mom_tracer_registry::tracer_type::flux_nameroot = "" Short tracer name snippet used construct the names of flux diagnostics.
-
character(len=64) mom_tracer_registry::tracer_type::flux_longname = "" A word or phrase used construct the long names of flux diagnostics.
-
real
flux_scale= 1.0¶ A scaling factor used to convert the fluxes of this tracer to its desired units.
-
character(len=48) mom_tracer_registry::tracer_type::flux_units = "" The units for fluxes of this variable.
-
character(len=48) mom_tracer_registry::tracer_type::conv_units = "" The units for the flux convergence of this tracer.
-
real
conv_scale= 1.0¶ A scaling factor used to convert the flux convergence of this tracer to its desired units.
-
character(len=48) mom_tracer_registry::tracer_type::cmor_tendprefix = "" The CMOR variable prefix for tendencies of this tracer, required because CMOR does not follow any discernable pattern for these names.
-
integer
ind_tr_squared= -1¶ The tracer registry index for the square of this tracer.
-
logical
advect_tr= .true.¶ If true, this tracer should be advected.
-
logical
hordiff_tr= .true.¶ If true, this tracer should experience epineutral diffusion.
-
logical
remap_tr= .true.¶ If true, this tracer should be vertically remapped.
-
integer
diag_form= 1¶ An integer indicating which template is to be used to label diagnostics.
-
integer
- type
A structure with diagnostic IDs of mass transport related diagnostics.
Unnamed Group
-
integer
id_uhtr= -1¶ Diagnostics for tracer horizontal transport.
-
integer
id_umo= -1¶ Diagnostics for tracer horizontal transport.
-
integer
id_umo_2d= -1¶ Diagnostics for tracer horizontal transport.
-
integer
id_vhtr= -1¶ Diagnostics for tracer horizontal transport.
-
integer
id_vmo= -1¶ Diagnostics for tracer horizontal transport.
-
integer
id_vmo_2d= -1¶ Diagnostics for tracer horizontal transport.
-
integer
id_dynamics_h= -1¶ Diagnostics for tracer horizontal transport.
-
integer
id_dynamics_h_tendency= -1¶ Diagnostics for tracer horizontal transport.
-
integer
-
interface
uchksum¶ Checksums an array (2d or 3d) staggered at C-grid u points.
Private Functions
-
subroutine
chksum_u_2d(array array, mesg mesg, HI HI, haloshift haloshift, symmetric symmetric, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums a 2d array staggered at C-grid u points.
- Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] mesg: An identifying message[in] haloshift: The width of halos to check (default 0)[in] symmetric: If true, do the checksums on the full symmetric computational domain.[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
chksum_u_3d(array array, mesg mesg, HI HI, haloshift haloshift, symmetric symmetric, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums a 3d array staggered at C-grid u points.
- Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] mesg: An identifying message[in] haloshift: The width of halos to check (default 0)[in] symmetric: If true, do the checksums on the full symmetric computational domain.[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
- type
Describes various unit conversion factors.
Public Members
-
real
m_to_z¶ A constant that translates distances in meters to the units of depth.
-
real
z_to_m¶ A constant that translates distances in the units of depth to meters.
-
real
m_to_l¶ A constant that translates lengths in meters to the units of horizontal lengths.
-
real
l_to_m¶ A constant that translates lengths in the units of horizontal lengths to meters.
-
real
s_to_t¶ A constant that time intervals in seconds to the units of time.
-
real
t_to_s¶ A constant that the units of time to seconds.
-
real
z_to_l¶ Convert vertical distances to lateral lengths.
-
real
l_to_z¶ Convert vertical distances to lateral lengths.
-
real
l_t_to_m_s¶ Convert lateral velocities from L T-1 to m s-1.
-
real
m_s_to_l_t¶ Convert lateral velocities from m s-1 to L T-1.
-
real
l_t2_to_m_s2¶ Convert lateral accelerations from L T-2 to m s-2.
-
real
z2_t_to_m2_s¶ Convert vertical diffusivities from Z2 T-1 to m2 s-1.
-
real
m2_s_to_z2_t¶ Convert vertical diffusivities from m2 s-1 to Z2 T-1.
-
real
m_to_z_restart= 0.0¶ A copy of the m_to_Z that is used in restart files.
-
real
m_to_l_restart= 0.0¶ A copy of the m_to_L that is used in restart files.
-
real
s_to_t_restart= 0.0¶ A copy of the s_to_T that is used in restart files.
-
real
- type
The control structure for the MOM_boundary_update module.
Unnamed Group
-
type(file_obc_cs), pointer mom_boundary_update::update_obc_cs::file_obc_csp => NULL() Pointers to the control structures for named OBC specifications.
-
type(kelvin_obc_cs), pointer mom_boundary_update::update_obc_cs::kelvin_obc_csp => NULL() Pointers to the control structures for named OBC specifications.
-
type(tidal_bay_obc_cs), pointer mom_boundary_update::update_obc_cs::tidal_bay_obc_csp => NULL() Pointers to the control structures for named OBC specifications.
-
type(shelfwave_obc_cs), pointer mom_boundary_update::update_obc_cs::shelfwave_obc_csp => NULL() Pointers to the control structures for named OBC specifications.
-
type(dyed_channel_obc_cs), pointer mom_boundary_update::update_obc_cs::dyed_channel_obc_csp => NULL() Pointers to the control structures for named OBC specifications.
Public Members
-
logical
use_files= .false.¶ If true, use external files for the open boundary.
-
logical
use_kelvin= .false.¶ If true, use the Kelvin wave open boundary.
-
logical
use_tidal_bay= .false.¶ If true, use the tidal_bay open boundary.
-
logical
use_shelfwave= .false.¶ If true, use the shelfwave open boundary.
-
logical
use_dyed_channel= .false.¶ If true, use the dyed channel open boundary.
-
- type
Control structure for user_change_diffusivity.
Public Members
-
real
kd_add¶ The scale of a diffusivity that is added everywhere without any filtering or scaling [Z2 T-1 ~> m2 s-1].
-
real, dimension(4) user_change_diffusivity::user_change_diff_cs::lat_range 4 values that define the latitude range over which a diffusivity scaled by Kd_add is added [degLat].
-
real, dimension(4) user_change_diffusivity::user_change_diff_cs::rho_range 4 values that define the coordinate potential density range over which a diffusivity scaled by Kd_add is added [kg m-3].
-
logical
use_abs_lat¶ If true, use the absolute value of latitude when setting lat_range.
-
type(diag_ctrl), pointer user_change_diffusivity::user_change_diff_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
real
- type
The control structure for the user_ice_shelf module.
Public Members
-
real
rho_ocean¶ The ocean’s typical density [kg m-2 Z-1].
-
real
max_draft¶ The maximum ocean draft of the ice shelf [Z ~> m].
-
real
min_draft¶ The minimum ocean draft of the ice shelf [Z ~> m].
-
real
flat_shelf_width¶ The range over which the shelf is min_draft thick [km].
-
real
shelf_slope_scale¶ The range over which the shelf slopes [km].
-
real
pos_shelf_edge_0¶ The x-position of the shelf edge at time 0 [km].
-
real
shelf_speed¶ The ice shelf speed of translation [km day-1].
-
logical
first_call= .true.¶ If true, this module has not been called before.
-
real
- type
Control structure for user_revise_forcing.
Public Members
-
real
cdrag¶ The quadratic bottom drag coefficient.
-
real
- type
This control structure should be used to store any run-time variables associated with the user-specified forcing.
It can be readily modified for a specific case, and because it is private there will be no changes needed in other code (although they will have to be recompiled).
Public Members
-
logical
use_temperature¶ If true, temperature and salinity are used as state variables.
-
logical
restorebuoy¶ If true, use restoring surface buoyancy forcing.
-
real
rho0¶ The density used in the Boussinesq approximation [kg m-3].
-
real
g_earth¶ The gravitational acceleration [L2 Z-1 s-2 ~> m s-2].
-
real
flux_const¶ The restoring rate at the surface [m s-1].
-
real
gust_const¶ A constant unresolved background gustiness that contributes to ustar [Pa].
-
type(diag_ctrl), pointer user_surface_forcing::user_surface_forcing_cs::diag A structure that is used to regulate the timing of diagnostic output.
-
logical
- type
The control structure for the USER_tracer_example module.
Public Members
-
logical
coupled_tracers= .false.¶ These tracers are not offered to the coupler.
-
character(len=200) user_tracer_example::user_tracer_example_cs::tracer_ic_file The full path to the IC file, or ” ” to initialize internally.
-
type(time_type), pointer user_tracer_example::user_tracer_example_cs::time => NULL() A pointer to the ocean model’s clock.
-
type(tracer_registry_type), pointer user_tracer_example::user_tracer_example_cs::tr_reg => NULL() A pointer to the tracer registry.
-
real, dimension(:,:,:,:), pointer user_tracer_example::user_tracer_example_cs::tr => NULL() The array of tracers used in this subroutine, in g m-3?
-
real, dimension(ntr) user_tracer_example::user_tracer_example_cs::land_val = -1.0 The value of tr that is used where land is masked out.
-
logical
use_sponge¶ If true, sponges may be applied somewhere in the domain.
-
integer, dimension(ntr) user_tracer_example::user_tracer_example_cs::ind_tr Indices returned by aof_set_coupler_flux if it is used and the surface tracer concentrations are to be provided to the coupler.
-
type(diag_ctrl), pointer user_tracer_example::user_tracer_example_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
type(vardesc), dimension(ntr) user_tracer_example::user_tracer_example_cs::tr_desc Descriptions of each of the tracers.
-
logical
-
interface
uvchksum¶ Checksums a pair velocity arrays (2d or 3d) staggered at C-grid locations.
Private Functions
-
subroutine
chksum_uv_2d(mesg mesg, arrayU arrayU, arrayV arrayV, HI HI, haloshift haloshift, symmetric symmetric, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums a pair of 2d velocity arrays staggered at C-grid locations.
- Parameters
[in] mesg: Identifying messages[in] hi: A horizontal index type[in] arrayu: The u-component array to be checksummed[in] arrayv: The v-component array to be checksummed[in] haloshift: The width of halos to check (default 0)[in] symmetric: If true, do the checksums on the full symmetric computational domain.[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for these arrays.[in] logunit: IO unit for checksum logging
-
subroutine
chksum_uv_3d(mesg mesg, arrayU arrayU, arrayV arrayV, HI HI, haloshift haloshift, symmetric symmetric, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums a pair of 3d velocity arrays staggered at C-grid locations.
- Parameters
[in] mesg: Identifying messages[in] hi: A horizontal index type[in] arrayu: The u-component array to be checksummed[in] arrayv: The v-component array to be checksummed[in] haloshift: The width of halos to check (default 0)[in] symmetric: If true, do the checksums on the full symmetric computational domain.[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for these arrays.[in] logunit: IO unit for checksum logging
-
subroutine
- type
Type for describing a variable, typically a tracer.
Public Members
-
character(len=64) mom_io::vardesc::name Variable name in a NetCDF file.
-
character(len=48) mom_io::vardesc::units Physical dimensions of the variable.
-
character(len=240) mom_io::vardesc::longname Long name of the variable.
-
character(len=8) mom_io::vardesc::hor_grid Horizontal grid: u, v, h, q, Cu, Cv, T, Bu, or 1.
-
character(len=8) mom_io::vardesc::z_grid Vertical grid: L, i, or 1.
-
character(len=8) mom_io::vardesc::t_grid Time description: s, p, or 1.
-
character(len=64) mom_io::vardesc::cmor_field_name CMOR name.
-
character(len=64) mom_io::vardesc::cmor_units CMOR physical dimensions of the variable.
-
character(len=240) mom_io::vardesc::cmor_longname CMOR long name of the variable.
-
real
conversion¶ for unit conversions, such as needed to convert from intensive to extensive
-
- type
Variable mixing coefficients.
Unnamed Group
-
integer
id_sn_u= -1¶ Diagnostic identifier.
-
integer
id_sn_v= -1¶ Diagnostic identifier.
-
integer
id_l2u= -1¶ Diagnostic identifier.
-
integer
id_l2v= -1¶ Diagnostic identifier.
-
integer
id_res_fn= -1¶ Diagnostic identifier.
-
integer
id_n2_u= -1¶ Diagnostic identifier.
-
integer
id_n2_v= -1¶ Diagnostic identifier.
-
integer
id_s2_u= -1¶ Diagnostic identifier.
-
integer
id_s2_v= -1¶ Diagnostic identifier.
-
integer
id_rd_dx= -1¶ Diagnostic identifier.
-
integer
id_kh_u_qg= -1¶ Diagnostic identifier.
-
integer
id_kh_v_qg= -1¶ Diagnostic identifier.
-
type(diag_ctrl), pointer mom_lateral_mixing_coeffs::varmix_cs::diag A structure that is used to regulate the timing of diagnostic output.
Public Members
-
logical
use_variable_mixing¶ If true, use the variable mixing.
-
logical
resoln_scaled_kh¶ If true, scale away the Laplacian viscosity when the deformation radius is well resolved.
-
logical
resoln_scaled_khth¶ If true, scale away the thickness diffusivity when the deformation radius is well resolved.
-
logical
resoln_scaled_khtr¶ If true, scale away the tracer diffusivity when the deformation radius is well resolved.
-
logical
interpolate_res_fn¶ If true, interpolate the resolution function to the velocity points from the thickness points; otherwise interpolate the wave speed and calculate the resolution function independently at each point.
-
logical
use_stored_slopes¶ If true, stores isopycnal slopes in this structure.
-
logical
resoln_use_ebt¶ If true, uses the equivalent barotropic wave speed instead of first baroclinic wave for calculating the resolution fn.
-
logical
khth_use_ebt_struct¶ If true, uses the equivalent barotropic structure as the vertical structure of thickness diffusivity.
-
logical
calculate_cg1¶ If true, calls wave_speed() to calculate the first baroclinic wave speed and populate CScg1. This parameter is set depending on other parameters.
-
logical
calculate_rd_dx¶ If true, calculates Rd/dx and populate CSRd_dx_h. This parameter is set depending on other parameters.
-
logical
calculate_res_fns¶ If true, calculate all the resolution factors. This parameter is set depending on other parameters.
-
logical
calculate_eady_growth_rate¶ If true, calculate all the Eady growth rate. This parameter is set depending on other parameters.
-
real, dimension(:,:), pointer mom_lateral_mixing_coeffs::varmix_cs::sn_u => NULL() S*N at u-points [s-1].
-
real, dimension(:,:), pointer mom_lateral_mixing_coeffs::varmix_cs::sn_v => NULL() S*N at v-points [s-1].
-
real, dimension(:,:), pointer mom_lateral_mixing_coeffs::varmix_cs::l2u => NULL() Length scale^2 at u-points [m2].
-
real, dimension(:,:), pointer mom_lateral_mixing_coeffs::varmix_cs::l2v => NULL() Length scale^2 at v-points [m2].
-
real, dimension(:,:), pointer mom_lateral_mixing_coeffs::varmix_cs::cg1 => NULL() The first baroclinic gravity wave speed [m s-1].
-
real, dimension(:,:), pointer mom_lateral_mixing_coeffs::varmix_cs::res_fn_h => NULL() Non-dimensional function of the ratio the first baroclinic.
-
real, dimension(:,:), pointer mom_lateral_mixing_coeffs::varmix_cs::res_fn_q => NULL() Non-dimensional function of the ratio the first baroclinic.
-
real, dimension(:,:), pointer mom_lateral_mixing_coeffs::varmix_cs::res_fn_u => NULL() Non-dimensional function of the ratio the first baroclinic.
-
real, dimension(:,:), pointer mom_lateral_mixing_coeffs::varmix_cs::res_fn_v => NULL() Non-dimensional function of the ratio the first baroclinic.
-
real, dimension(:,:), pointer mom_lateral_mixing_coeffs::varmix_cs::beta_dx2_h => NULL() The magnitude of the gradient of the Coriolis parameter.
-
real, dimension(:,:), pointer mom_lateral_mixing_coeffs::varmix_cs::beta_dx2_q => NULL() The magnitude of the gradient of the Coriolis parameter.
-
real, dimension(:,:), pointer mom_lateral_mixing_coeffs::varmix_cs::beta_dx2_u => NULL() The magnitude of the gradient of the Coriolis parameter.
-
real, dimension(:,:), pointer mom_lateral_mixing_coeffs::varmix_cs::beta_dx2_v => NULL() The magnitude of the gradient of the Coriolis parameter.
-
real, dimension(:,:), pointer mom_lateral_mixing_coeffs::varmix_cs::f2_dx2_h => NULL() The Coriolis parameter squared times the grid.
-
real, dimension(:,:), pointer mom_lateral_mixing_coeffs::varmix_cs::f2_dx2_q => NULL() The Coriolis parameter squared times the grid.
-
real, dimension(:,:), pointer mom_lateral_mixing_coeffs::varmix_cs::f2_dx2_u => NULL() The Coriolis parameter squared times the grid.
-
real, dimension(:,:), pointer mom_lateral_mixing_coeffs::varmix_cs::f2_dx2_v => NULL() The Coriolis parameter squared times the grid.
-
real, dimension(:,:), pointer mom_lateral_mixing_coeffs::varmix_cs::rd_dx_h => NULL() Deformation radius over grid spacing [nondim].
-
real, dimension(:,:,:), pointer mom_lateral_mixing_coeffs::varmix_cs::slope_x => NULL() Zonal isopycnal slope [nondim].
-
real, dimension(:,:,:), pointer mom_lateral_mixing_coeffs::varmix_cs::slope_y => NULL() Meridional isopycnal slope [nondim].
-
real, dimension(:,:,:), pointer mom_lateral_mixing_coeffs::varmix_cs::n2_u => NULL() Brunt-Vaisala frequency at u-points [s-2].
-
real, dimension(:,:,:), pointer mom_lateral_mixing_coeffs::varmix_cs::n2_v => NULL() Brunt-Vaisala frequency at v-points [s-2].
-
real, dimension(:,:,:), pointer mom_lateral_mixing_coeffs::varmix_cs::ebt_struct => NULL() Vertical structure function to scale diffusivities with [nondim].
-
real, dimension( : , : ), allocatable mom_lateral_mixing_coeffs::varmix_cs::laplac3_const_u Laplacian metric-dependent constants [nondim].
-
real, dimension( : , : ), allocatable mom_lateral_mixing_coeffs::varmix_cs::laplac3_const_v Laplacian metric-dependent constants [nondim].
-
real, dimension( : , : , : ), allocatable mom_lateral_mixing_coeffs::varmix_cs::kh_u_qg QG Leith GM coefficient at u-points [m2 s-1].
-
real, dimension( : , : , : ), allocatable mom_lateral_mixing_coeffs::varmix_cs::kh_v_qg QG Leith GM coefficient at v-points [m2 s-1].
-
logical
use_visbeck¶ Use Visbeck formulation for thickness diffusivity.
-
integer
varmix_ktop¶ Top layer to start downward integrals.
-
real
visbeck_l_scale¶ Fixed length scale in Visbeck formula.
-
real
res_coef_khth¶ A non-dimensional number that determines the function of resolution, used for thickness and tracer mixing, as: F = 1 / (1 + (Res_coef_khth*Ld/dx)^Res_fn_power)
-
real
res_coef_visc¶ A non-dimensional number that determines the function of resolution, used for lateral viscosity, as: F = 1 / (1 + (Res_coef_visc*Ld/dx)^Res_fn_power)
-
real
kappa_smooth¶ A diffusivity for smoothing T/S in vanished layers [m2 s-1].
-
integer
res_fn_power_khth¶ The power of dx/Ld in the KhTh resolution function. Any positive integer power may be used, but even powers and especially 2 are coded to be more efficient.
-
integer
res_fn_power_visc¶ The power of dx/Ld in the Kh resolution function. Any positive integer power may be used, but even powers and especially 2 are coded to be more efficient.
-
real
visbeck_s_max¶ Upper bound on slope used in Eady growth rate [nondim].
-
logical
use_qg_leith_gm¶ If true, uses the QG Leith viscosity as the GM coefficient.
-
logical
use_beta_in_qg_leith¶ If true, includes the beta term in the QG Leith GM coefficient.
-
type(wave_speed_cs), pointer mom_lateral_mixing_coeffs::varmix_cs::wave_speed_csp => NULL() Wave speed control structure.
-
type(group_pass_type) mom_lateral_mixing_coeffs::varmix_cs::pass_cg1 For group halo pass.
-
logical
debug¶ If true, write out checksums of data for debugging.
-
integer
-
interface
vchksum¶ Checksums an array (2d or 3d) staggered at C-grid v points.
Private Functions
-
subroutine
chksum_v_2d(array array, mesg mesg, HI HI, haloshift haloshift, symmetric symmetric, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums a 2d array staggered at C-grid v points.
- Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] mesg: An identifying message[in] haloshift: The width of halos to check (default 0)[in] symmetric: If true, do the checksums on the full symmetric computational domain.[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
chksum_v_3d(array array, mesg mesg, HI HI, haloshift haloshift, symmetric symmetric, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums a 3d array staggered at C-grid v points.
- Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] mesg: An identifying message[in] haloshift: The width of halos to check (default 0)[in] symmetric: If true, do the checksums on the full symmetric computational domain.[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
-
interface
vec_chksum¶ Do checksums on the components of a C-grid vector.
Private Functions
-
subroutine
chksum_vec_c3d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, halos halos, scalars scalars)¶ Do a checksum and redundant point check on a 3d C-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector[in] v_comp: The v-component of the vector[in] halos: The width of halos to check (default 0)[in] scalars: If true this is a pair of scalars that are being checked.
-
subroutine
chksum_vec_c2d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, halos halos, scalars scalars)¶ Do a checksum and redundant point check on a 2d C-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector[in] v_comp: The v-component of the vector[in] halos: The width of halos to check (default 0)[in] scalars: If true this is a pair of scalars that are being checked.
-
subroutine
-
interface
vec_chksum_a¶ Do checksums on the components of an A-grid vector.
Private Functions
-
subroutine
chksum_vec_a3d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, halos halos, scalars scalars)¶ Do a checksum and redundant point check on a 3d C-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector[in] v_comp: The v-component of the vector[in] halos: The width of halos to check (default 0)[in] scalars: If true this is a pair of scalars that are being checked.
-
subroutine
chksum_vec_a2d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, halos halos, scalars scalars)¶ Do a checksum and redundant point check on a 2d C-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector[in] v_comp: The v-component of the vector[in] halos: The width of halos to check (default 0)[in] scalars: If true this is a pair of scalars that are being checked.
-
subroutine
-
interface
vec_chksum_b¶ Do checksums on the components of a B-grid vector.
Private Functions
-
subroutine
chksum_vec_b3d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, halos halos, scalars scalars)¶ Do a checksum and redundant point check on a 3d B-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector[in] v_comp: The v-component of the vector[in] halos: The width of halos to check (default 0)[in] scalars: If true this is a pair of scalars that are being checked.
-
subroutine
chksum_vec_b2d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, halos halos, scalars scalars, symmetric symmetric)¶ - Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector[in] v_comp: The v-component of the vector[in] halos: The width of halos to check (default 0)[in] scalars: If true this is a pair of scalars that are being checked.[in] symmetric: If true, do the checksums on the full symmetric computational domain.
-
subroutine
-
interface
vec_chksum_c¶ Do checksums on the components of a C-grid vector.
Private Functions
-
subroutine
chksum_vec_c3d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, halos halos, scalars scalars)¶ Do a checksum and redundant point check on a 3d C-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector[in] v_comp: The v-component of the vector[in] halos: The width of halos to check (default 0)[in] scalars: If true this is a pair of scalars that are being checked.
-
subroutine
chksum_vec_c2d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, halos halos, scalars scalars)¶ Do a checksum and redundant point check on a 2d C-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector[in] v_comp: The v-component of the vector[in] halos: The width of halos to check (default 0)[in] scalars: If true this is a pair of scalars that are being checked.
-
subroutine
- type
Describes the vertical ocean grid, including unit conversion factors.
Public Members
-
integer
ke¶ The number of layers/levels in the vertical.
-
real
max_depth¶ The maximum depth of the ocean [Z ~> m].
-
real
mks_g_earth¶ The gravitational acceleration in unscaled MKS units [m s-2].
-
real
g_earth¶ The gravitational acceleration [L2 Z-1 T-2 ~> m s-2].
-
real
rho0¶ The density used in the Boussinesq approximation or nominal density used to convert depths into mass units [kg m-3].
-
character(len=40) mom_verticalgrid::verticalgrid_type::zaxisunits The units that vertical coordinates are written in.
-
character(len=40) mom_verticalgrid::verticalgrid_type::zaxislongname Coordinate name to appear in files, e.g. “Target Potential Density” or “Height”.
-
real, dimension(:), allocatable mom_verticalgrid::verticalgrid_type::slayer Coordinate values of layer centers.
-
real, dimension(:), allocatable mom_verticalgrid::verticalgrid_type::sinterface Coordinate values on interfaces.
-
integer
direction= 1¶ Direction defaults to 1, positive up.
-
logical
boussinesq¶ If true, make the Boussinesq approximation.
-
real
angstrom_h¶ A one-Angstrom thickness in the model thickness units [H ~> m or kg m-2].
-
real
angstrom_z¶ A one-Angstrom thickness in the model depth units [Z ~> m].
-
real
angstrom_m¶ A one-Angstrom thickness [m].
-
real
h_subroundoff¶ A thickness that is so small that it can be added to a thickness of Angstrom or larger without changing it at the bit level [H ~> m or kg m-2]. If Angstrom is 0 or exceedingly small, this is negligible compared to 1e-17 m.
-
real, dimension(:), allocatable mom_verticalgrid::verticalgrid_type::g_prime The reduced gravity at each interface [L2 Z-1 T-2 ~> m s-2].
-
real, dimension(:), allocatable mom_verticalgrid::verticalgrid_type::rlay The target coordinate value (potential density) in each layer [kg m-3].
-
integer
nkml= 0¶ The number of layers at the top that should be treated as parts of a homogeneous region.
-
integer
nk_rho_varies= 0¶ The number of layers at the top where the density does not track any target density.
-
real
h_to_kg_m2¶ A constant that translates thicknesses from the units of thickness to kg m-2.
-
real
kg_m2_to_h¶ A constant that translates thicknesses from kg m-2 to the units of thickness.
-
real
m_to_h¶ A constant that translates distances in m to the units of thickness.
-
real
h_to_m¶ A constant that translates distances in the units of thickness to m.
-
real
h_to_pa¶ A constant that translates the units of thickness to pressure [Pa].
-
real
h_to_z¶ A constant that translates thickness units to the units of depth.
-
real
z_to_h¶ A constant that translates depth units to thickness units.
-
real
m_to_h_restart= 0.0¶ A copy of the m_to_H that is used in restart files.
-
integer
- type
The control structure with parameters and memory for the MOM_vert_friction module.
Unnamed Group
-
integer
id_du_dt_visc= -1¶ Diagnostic identifiers.
-
integer
id_dv_dt_visc= -1¶ Diagnostic identifiers.
-
integer
id_au_vv= -1¶ Diagnostic identifiers.
-
integer
id_av_vv= -1¶ Diagnostic identifiers.
-
integer
id_h_u= -1¶ Diagnostic identifiers.
-
integer
id_h_v= -1¶ Diagnostic identifiers.
-
integer
id_hml_u= -1¶ Diagnostic identifiers.
-
integer
id_hml_v= -1¶ Diagnostic identifiers.
-
integer
id_ray_u= -1¶ Diagnostic identifiers.
-
integer
id_ray_v= -1¶ Diagnostic identifiers.
-
integer
id_taux_bot= -1¶ Diagnostic identifiers.
-
integer
id_tauy_bot= -1¶ Diagnostic identifiers.
-
integer
id_kv_slow= -1¶ Diagnostic identifiers.
-
integer
id_kv_u= -1¶ Diagnostic identifiers.
-
integer
id_kv_v= -1¶ Diagnostic identifiers.
Public Members
-
real
hmix¶ The mixed layer thickness in thickness units [H ~> m or kg m-2].
-
real
hmix_stress¶ The mixed layer thickness over which the wind stress is applied with direct_stress [H ~> m or kg m-2].
-
real
kvml¶ The mixed layer vertical viscosity [Z2 T-1 ~> m2 s-1].
-
real
kv¶ The interior vertical viscosity [Z2 T-1 ~> m2 s-1].
-
real
hbbl¶ The static bottom boundary layer thickness [H ~> m or kg m-2].
-
real
kvbbl¶ The vertical viscosity in the bottom boundary layer [Z2 T-1 ~> m2 s-1].
-
real
maxvel¶ Velocity components greater than maxvel are truncated [m s-1].
-
real
vel_underflow¶ Velocity components smaller than vel_underflow are set to 0 [m s-1].
-
logical
cfl_based_trunc¶ If true, base truncations on CFL numbers, not absolute velocities.
-
real
cfl_trunc¶ Velocity components will be truncated when they are large enough that the corresponding CFL number exceeds this value, nondim.
-
real
cfl_report¶ The value of the CFL number that will cause the accelerations to be reported, nondim. CFL_report will often equal CFL_trunc.
-
real
truncramptime¶ The time-scale over which to ramp up the value of CFL_trunc from CFL_truncS to CFL_truncE.
-
real
cfl_truncs¶ The start value of CFL_trunc.
-
real
cfl_trunce¶ The end/target value of CFL_trunc.
-
logical
cflrampingisactivated= .false.¶ True if the ramping has been initialized.
-
type(time_type) mom_vert_friction::vertvisc_cs::rampstarttime The time at which the ramping of CFL_trunc starts.
-
real, dimension( : , : , : ), allocatable mom_vert_friction::vertvisc_cs::a_u The u-drag coefficient across an interface [Z T-1 ~> m s-1].
-
real, dimension( : , : , : ), allocatable mom_vert_friction::vertvisc_cs::h_u The effective layer thickness at u-points [H ~> m or kg m-2].
-
real, dimension( : , : , : ), allocatable mom_vert_friction::vertvisc_cs::a_v The v-drag coefficient across an interface [Z T-1 ~> m s-1].
-
real, dimension( : , : , : ), allocatable mom_vert_friction::vertvisc_cs::h_v The effective layer thickness at v-points [H ~> m or kg m-2].
-
real, dimension(:,:), pointer mom_vert_friction::vertvisc_cs::a1_shelf_u => NULL() The u-momentum coupling coefficient under ice shelves [Z T-1 ~> m s-1]. Retained to determine stress under shelves.
-
real, dimension(:,:), pointer mom_vert_friction::vertvisc_cs::a1_shelf_v => NULL() The v-momentum coupling coefficient under ice shelves [Z T-1 ~> m s-1]. Retained to determine stress under shelves.
-
logical
split¶ If true, use the split time stepping scheme.
-
logical
bottomdraglaw¶ If true, the bottom stress is calculated with a drag law c_drag*|u|*u. The velocity magnitude may be an assumed value or it may be based on the actual velocity in the bottommost HBBL, depending on whether linear_drag is true.
-
logical
channel_drag¶ If true, the drag is exerted directly on each layer according to what fraction of the bottom they overlie.
-
logical
harmonic_visc¶ If true, the harmonic mean thicknesses are used to calculate the viscous coupling between layers except near the bottom. Otherwise the arithmetic mean thickness is used except near the bottom.
-
real
harm_bl_val¶ A scale to determine when water is in the boundary layers based solely on harmonic mean thicknesses for the purpose of determining the extent to which the thicknesses used in the viscosities are upwinded.
-
logical
direct_stress¶ If true, the wind stress is distributed over the topmost Hmix_stress of fluid and KVML may be very small.
-
logical
dynamic_viscous_ml¶ If true, use the results from a dynamic calculation, perhaps based on a bulk Richardson number criterion, to determine the mixed layer thickness for viscosity.
-
logical
debug¶ If true, write verbose checksums for debugging purposes.
-
integer
nkml¶ The number of layers in the mixed layer.
-
integer, pointer mom_vert_friction::vertvisc_cs::ntrunc The number of times the velocity has been truncated since the last call to write_energy.
-
character(len=200) mom_vert_friction::vertvisc_cs::u_trunc_file The complete path to a file in which a column of u-accelerations are written if velocity truncations occur.
-
character(len=200) mom_vert_friction::vertvisc_cs::v_trunc_file The complete path to a file in which a column of v-accelerations are written if velocity truncations occur.
-
logical
stokesmixing¶ If true, do Stokes drift mixing via the Lagrangian current (Eulerian plus Stokes drift). False by default and set via STOKES_MIXING_COMBINED.
-
type(diag_ctrl), pointer mom_vert_friction::vertvisc_cs::diag A structure that is used to regulate the timing of diagnostic output.
-
type(pointaccel_cs), pointer mom_vert_friction::vertvisc_cs::pointaccel_csp => NULL() A pointer to the control structure for recording accelerations leading to velocity truncations.
-
integer
- type
Vertical viscosities, drag coefficients, and related fields.
Public Members
-
real
prandtl_turb¶ The Prandtl number for the turbulent diffusion that is captured in Kd_shear [nondim].
-
real, dimension(:,:), pointer mom_variables::vertvisc_type::bbl_thick_u => NULL() The bottom boundary layer thickness at the u-points [Z ~> m].
-
real, dimension(:,:), pointer mom_variables::vertvisc_type::bbl_thick_v => NULL() The bottom boundary layer thickness at the v-points [Z ~> m].
-
real, dimension(:,:), pointer mom_variables::vertvisc_type::kv_bbl_u => NULL() The bottom boundary layer viscosity at the u-points [Z2 T-1 ~> m2 s-1].
-
real, dimension(:,:), pointer mom_variables::vertvisc_type::kv_bbl_v => NULL() The bottom boundary layer viscosity at the v-points [Z2 T-1 ~> m2 s-1].
-
real, dimension(:,:), pointer mom_variables::vertvisc_type::ustar_bbl => NULL() The turbulence velocity in the bottom boundary layer at h points [Z T-1 ~> m s-1].
-
real, dimension(:,:), pointer mom_variables::vertvisc_type::tke_bbl => NULL() A term related to the bottom boundary layer source of turbulent kinetic energy, currently in [Z3 T-3 ~> m3 s-3], but may at some time be changed to [kg Z3 m-3 T-3 ~> W m-2].
-
real, dimension(:,:), pointer mom_variables::vertvisc_type::taux_shelf => NULL() The zonal stresses on the ocean under shelves [Pa].
-
real, dimension(:,:), pointer mom_variables::vertvisc_type::tauy_shelf => NULL() The meridional stresses on the ocean under shelves [Pa].
-
real, dimension(:,:), pointer mom_variables::vertvisc_type::tbl_thick_shelf_u => NULL() Thickness of the viscous top boundary layer under ice shelves at u-points [Z ~> m].
-
real, dimension(:,:), pointer mom_variables::vertvisc_type::tbl_thick_shelf_v => NULL() Thickness of the viscous top boundary layer under ice shelves at v-points [Z ~> m].
-
real, dimension(:,:), pointer mom_variables::vertvisc_type::kv_tbl_shelf_u => NULL() Viscosity in the viscous top boundary layer under ice shelves at u-points [Z2 T-1 ~> m2 s-1].
-
real, dimension(:,:), pointer mom_variables::vertvisc_type::kv_tbl_shelf_v => NULL() Viscosity in the viscous top boundary layer under ice shelves at v-points [Z2 T-1 ~> m2 s-1].
-
real, dimension(:,:), pointer mom_variables::vertvisc_type::nkml_visc_u => NULL() The number of layers in the viscous surface mixed layer at u-points [nondim]. This is not an integer because there may be fractional layers, and it is stored in terms of layers, not depth, to facilitate the movement of the viscous boundary layer with the flow.
-
real, dimension(:,:), pointer mom_variables::vertvisc_type::nkml_visc_v => NULL() The number of layers in the viscous surface mixed layer at v-points [nondim].
-
real, dimension(:,:), pointer mom_variables::vertvisc_type::mld => NULL() Instantaneous active mixing layer depth [H ~> m or kg m-2].
-
real, dimension(:,:,:), pointer mom_variables::vertvisc_type::ray_u => NULL() The Rayleigh drag velocity to be applied to each layer at u-points [Z T-1 ~> m s-1].
-
real, dimension(:,:,:), pointer mom_variables::vertvisc_type::ray_v => NULL() The Rayleigh drag velocity to be applied to each layer at v-points [Z T-1 ~> m s-1].
-
real, dimension(:,:,:), pointer mom_variables::vertvisc_type::kd_extra_t => NULL() The extra diffusivity of temperature due to double diffusion relative to the diffusivity of density [Z2 T-1 ~> m2 s-1].
-
real, dimension(:,:,:), pointer mom_variables::vertvisc_type::kd_extra_s => NULL() The extra diffusivity of salinity due to double diffusion relative to the diffusivity of density [Z2 T-1 ~> m2 s-1].
-
real, dimension(:,:,:), pointer mom_variables::vertvisc_type::kd_shear => NULL() The shear-driven turbulent diapycnal diffusivity at the interfaces between layers in tracer columns [Z2 T-1 ~> m2 s-1].
-
real, dimension(:,:,:), pointer mom_variables::vertvisc_type::kv_shear => NULL() The shear-driven turbulent vertical viscosity at the interfaces between layers in tracer columns [Z2 T-1 ~> m2 s-1].
-
real, dimension(:,:,:), pointer mom_variables::vertvisc_type::kv_shear_bu => NULL() The shear-driven turbulent vertical viscosity at the interfaces between layers in corner columns [Z2 T-1 ~> m2 s-1].
-
real, dimension(:,:,:), pointer mom_variables::vertvisc_type::kv_slow => NULL() The turbulent vertical viscosity component due to “slow” processes (e.g., tidal, background, convection etc) [Z2 T-1 ~> m2 s-1].
-
real, dimension(:,:,:), pointer mom_variables::vertvisc_type::tke_turb => NULL() The turbulent kinetic energy per unit mass at the interfaces [Z2 T-2 ~> m2 s-2]. This may be at the tracer or corner points.
-
logical
add_kv_slow¶ If True, add Kv_slow when calculating the ‘coupling coefficient’ (a_cpl) at the interfaces in find_coupling_coef.
-
real
- type
Container for all surface wave related parameters.
Unnamed Group
-
integer, public mom_wave_interface::wave_parameters_cs::id_surfacestokes_x = -1 Diagnostic handles.
-
integer, public mom_wave_interface::wave_parameters_cs::id_surfacestokes_y = -1 Diagnostic handles.
-
integer, public mom_wave_interface::wave_parameters_cs::id_3dstokes_x = -1 Diagnostic handles.
-
integer, public mom_wave_interface::wave_parameters_cs::id_3dstokes_y = -1 Diagnostic handles.
-
integer, public mom_wave_interface::wave_parameters_cs::id_la_turb = -1 Diagnostic handles.
Public Members
-
logical, public mom_wave_interface::wave_parameters_cs::usewaves Flag to enable surface gravity wave feature.
-
logical, public mom_wave_interface::wave_parameters_cs::lagrangianmixing This feature is in development and not ready True if Stokes drift is present and mixing should be applied to Lagrangian current (mean current + Stokes drift). See Reichl et al., 2016 KPP-LT approach.
-
logical, public mom_wave_interface::wave_parameters_cs::stokesmixing This feature is in development and not ready. True if vertical mixing of momentum should be applied directly to Stokes current (with separate mixing parameter for Eulerian mixing contribution). See Harcourt 2013, 2015 Second-Moment approach.
-
logical, public mom_wave_interface::wave_parameters_cs::coriolisstokes This feature is in development and not ready.
-
integer, public mom_wave_interface::wave_parameters_cs::stklevelmode =1 Sets if Stokes drift is defined at mid-points or layer averaged. Set to 0 if mid-point and set to 1 if average value of Stokes drift over level. If advecting with Stokes transport, 1 is the correct approach.
-
real, dimension(:), allocatable, public mom_wave_interface::wave_parameters_cs::wavenum_cen Wavenumber bands for read/coupled [m-1].
-
real, dimension(:), allocatable, public mom_wave_interface::wave_parameters_cs::freq_cen Frequency bands for read/coupled [s-1].
-
real, dimension(:), allocatable, public mom_wave_interface::wave_parameters_cs::prescribedsurfstkx Surface Stokes drift if prescribed [m s-1].
-
real, dimension(:), allocatable, public mom_wave_interface::wave_parameters_cs::prescribedsurfstky Surface Stokes drift if prescribed [m s-1].
-
real, dimension(:,:,:), allocatable, public mom_wave_interface::wave_parameters_cs::us_x 3d zonal Stokes drift profile [m s-1]
-
real, dimension(:,:,:), allocatable, public mom_wave_interface::wave_parameters_cs::us_y 3d meridional Stokes drift profile [m s-1]
-
real, dimension(:,:), allocatable, public mom_wave_interface::wave_parameters_cs::la_sl SL Langmuir number (directionality factored later)
-
real, dimension(:,:), allocatable, public mom_wave_interface::wave_parameters_cs::la_turb Aligned Turbulent Langmuir number.
-
real, dimension(:,:), allocatable, public mom_wave_interface::wave_parameters_cs::us0_x Surface Stokes Drift (zonal, m/s)
-
real, dimension(:,:), allocatable, public mom_wave_interface::wave_parameters_cs::us0_y Surface Stokes Drift (meridional, m/s)
-
real, dimension(:,:,:), allocatable, public mom_wave_interface::wave_parameters_cs::stkx0 Stokes Drift spectrum (zonal, m/s)
-
real, dimension(:,:,:), allocatable, public mom_wave_interface::wave_parameters_cs::stky0 Stokes Drift spectrum (meridional, m/s)
-
real, dimension(:,:,:), allocatable, public mom_wave_interface::wave_parameters_cs::kvs Viscosity for Stokes Drift shear [Z2 T-1 ~> m2 s-1].
-
type(time_type), pointer, public mom_wave_interface::wave_parameters_cs::time A pointer to the ocean model’s clock.
-
type(diag_ctrl), pointer, public mom_wave_interface::wave_parameters_cs::diag A structure that is used to regulate the timing of diagnostic output.
-
real
la_min= 0.05¶ An arbitrary lower-bound on the Langmuir number. Run-time parameter. Langmuir number is sqrt(u_star/u_stokes). When both are small but u_star is orders of magnitude smaller the Langmuir number could have unintended consequences. Since both are small it can be safely capped to avoid such consequences.
-
- type
Control structure for MOM_wave_speed.
Public Members
-
logical
use_ebt_mode= .false.¶ If true, calculate the equivalent barotropic wave speed instead of the first baroclinic wave speed. This parameter controls the default behavior of wave_speed() which can be overridden by optional arguments.
-
real
mono_n2_column_fraction= 0.¶ The lower fraction of water column over which N2 is limited as monotonic for the purposes of calculating the equivalent barotropic wave speed. This parameter controls the default behavior of wave_speed() which can be overridden by optional arguments.
-
real
mono_n2_depth= -1.¶ The depth below which N2 is limited as monotonic for the purposes of calculating the equivalent barotropic wave speed [Z ~> m]. This parameter controls the default behavior of wave_speed() which can be overridden by optional arguments.
-
type(remapping_cs) mom_wave_speed::wave_speed_cs::remapping_cs Used for vertical remapping when calculating equivalent barotropic mode structure.
-
type(diag_ctrl), pointer mom_wave_speed::wave_speed_cs::diag Diagnostics control structure.
-
logical
- type
The control structure for the MOM_wave_structure module.
Public Members
-
type(diag_ctrl), pointer mom_wave_structure::wave_structure_cs::diag => NULL() A structure that is used to regulate the timing of diagnostic output.
-
real, dimension(:,:,:), allocatable mom_wave_structure::wave_structure_cs::w_strct Vertical structure of vertical velocity (normalized) [m s-1].
-
real, dimension(:,:,:), allocatable mom_wave_structure::wave_structure_cs::u_strct Vertical structure of horizontal velocity (normalized) [m s-1].
-
real, dimension(:,:,:), allocatable mom_wave_structure::wave_structure_cs::w_profile Vertical profile of w_hat(z), where w(x,y,z,t) = w_hat(z)*exp(i(kx+ly-freq*t)) is the full time- varying vertical velocity with w_hat(z) = W0*w_strct(z) [m s-1].
-
real, dimension(:,:,:), allocatable mom_wave_structure::wave_structure_cs::uavg_profile Vertical profile of the magnitude of horizontal velocity, (u^2+v^2)^0.5, averaged over a period [m s-1].
-
real, dimension(:,:,:), allocatable mom_wave_structure::wave_structure_cs::z_depths Depths of layer interfaces [m].
-
real, dimension(:,:,:), allocatable mom_wave_structure::wave_structure_cs::n2 Squared buoyancy frequency at each interface [s-2].
-
integer, dimension(:,:), allocatable mom_wave_structure::wave_structure_cs::num_intfaces Number of layer interfaces (including surface and bottom)
-
real
int_tide_source_x¶ X Location of generation site for internal tide for testing (BDM)
-
real
int_tide_source_y¶ Y Location of generation site for internal tide for testing (BDM)
-
- type
A control structure that regulates the writing of CPU time.
Public Members
-
real
maxcpu¶ The maximum amount of cpu time per processor for which MOM should run before saving a restart file and quiting with a return value that indicates that further execution is required to complete the simulation, in wall-clock seconds.
-
type(time_type) mom_write_cputime::write_cputime_cs::start_time The start time of the simulation. Start_time is set in MOM_initialization.F90.
-
real
startup_cputime¶ The CPU time used in the startup phase of the model.
-
real
prev_cputime= 0.0¶ The last measured CPU time.
-
real
dn_dcpu_min= -1.0¶ The minimum derivative of timestep with CPU time.
-
real
cputime2= 0.0¶ The accumulated cpu time.
-
integer
previous_calls= 0¶ The number of times write_CPUtime has been called.
-
integer
prev_n= 0¶ The value of n from the last call.
-
integer
filecpu_ascii¶ The unit number of the CPU time file.
-
character(len=200) mom_write_cputime::write_cputime_cs::cpufile The name of the CPU time file.
-
real
- type
Control structure containing required parameters for a z-like coordinate.
Public Members
-
integer
nk¶ Number of levels to be generated.
-
real
min_thickness¶ Minimum thickness allowed for layers, in the same thickness units (perhaps [H ~> m or kg m-2]) that will be used in all subsequent calls to build_zstar_column with this structure.
-
real, dimension(:), allocatable coord_zlike::zlike_cs::coordinateresolution Target coordinate resolution, usually in [Z ~> m].
-
integer
-
namespace
adjustment_initialization¶ Configures the model for the geostrophic adjustment test case.
Functions
-
subroutine, public adjustment_initialization::adjustment_initialize_thickness(h h, G G, GV GV, US US, param_file param_file, just_read_params just_read_params) Initializes the layer thicknesses in the adjustment test case.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[out] h: The thickness that is being initialized [H ~> m or kg m-2].[in] param_file: A structure indicating the open file to parse for model parameter values.[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine, public adjustment_initialization::adjustment_initialize_temperature_salinity(T T, S S, h h, G G, GV GV, param_file param_file, eqn_of_state eqn_of_state, just_read_params just_read_params) Initialization of temperature and salinity in the adjustment test case.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[out] t: The temperature that is being initialized.[out] s: The salinity that is being initialized.[in] h: The model thicknesses [H ~> m or kg m-2].[in] param_file: A structure indicating the open file to parse for model parameter values.eqn_of_state: Equation of state.[in] just_read_params: If present and true, this call will only read parameters without changing T & S.
Variables
-
character(len=40) adjustment_initialization::mdl = "adjustment_initialization" This module’s name.
-
-
namespace
advection_test_tracer¶ This tracer package is used to test advection schemes.
Functions
-
logical function, public advection_test_tracer::register_advection_test_tracer(HI HI, GV GV, param_file param_file, CS CS, tr_Reg tr_Reg, restart_CS restart_CS) Register tracer fields and subroutines to be used with MOM.
- Parameters
[in] hi: A horizontal index type structure[in] gv: The ocean’s vertical grid structure[in] param_file: A structure to parse for run-time parameterscs: The control structure returned by a previous call to register_advection_test_tracer.tr_reg: A pointer that is set to point to the control structure for the tracer advection and diffusion modulerestart_cs: A pointer to the restart control structure
-
subroutine, public advection_test_tracer::initialize_advection_test_tracer(restart restart, day day, G G, GV GV, h h, diag diag, OBC OBC, CS CS, sponge_CSp sponge_CSp) Initializes the NTR tracer fields in tr(:,:,:,:) and it sets up the tracer output.
- Parameters
[in] restart: .true. if the fields have already been read from a restart file.[in] day: Time of the start of the run.[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][in] diag: A structure that is used to regulate diagnostic output.obc: This open boundary condition type specifies whether, where, and what open boundary conditions are used.cs: The control structure returned by a previous call to register_advection_test_tracer.sponge_csp: Pointer to the control structure for the sponges.
-
subroutine, public advection_test_tracer::advection_test_tracer_column_physics(h_old h_old, h_new h_new, ea ea, eb eb, fluxes fluxes, dt dt, G G, GV GV, CS CS, evap_CFL_limit evap_CFL_limit, minimum_forcing_depth minimum_forcing_depth) Applies diapycnal diffusion and any other column tracer physics or chemistry to the tracers from this package. This is a simple example of a set of advected passive tracers.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h_old: Layer thickness before entrainment [H ~> m or kg m-2].[in] h_new: Layer thickness after entrainment [H ~> m or kg m-2].[in] ea: an array to which the amount of fluid entrained[in] eb: an array to which the amount of fluid entrained[in] fluxes: A structure containing pointers to thermodynamic and tracer forcing fields. Unused fields have NULL ptrs.[in] dt: The amount of time covered by this call [s]cs: The control structure returned by a previous call to register_advection_test_tracer.[in] evap_cfl_limit: Limit on the fraction of the water that can be fluxed out of the top layer in a timestep [nondim][in] minimum_forcing_depth: The smallest depth over which fluxes can be applied [m]
-
subroutine, public advection_test_tracer::advection_test_tracer_surface_state(state state, h h, G G, CS CS) This subroutine extracts the surface fields from this tracer package that are to be shared with the atmosphere in coupled configurations. This particular tracer package does not report anything back to the coupler.
- Parameters
[in] g: The ocean’s grid structure.[inout] state: A structure containing fields that describe the surface state of the ocean.[in] h: Layer thickness [H ~> m or kg m-2].cs: The control structure returned by a previous call to register_advection_test_tracer.
-
integer function, public advection_test_tracer::advection_test_stock(h h, stocks stocks, G G, GV GV, CS CS, names names, units units, stock_index stock_index) Calculate the mass-weighted integral of all tracer stocks, returning the number of stocks it has calculated. If the stock_index is present, only the stock corresponding to that coded index is returned.
- Return
the number of stocks calculated here.
- Parameters
[in] g: The ocean’s grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][out] stocks: the mass-weighted integrated amount of each tracer, in kg times concentration units [kg conc].[in] gv: The ocean’s vertical grid structurecs: The control structure returned by a previous call to register_advection_test_tracer.[out] names: the names of the stocks calculated.[out] units: the units of the stocks calculated.[in] stock_index: the coded index of a specific stock being sought.
-
subroutine, public advection_test_tracer::advection_test_tracer_end(CS CS) Deallocate memory associated with this module.
- Parameters
cs: The control structure returned by a previous call to register_advection_test_tracer.
Variables
-
integer, parameter advection_test_tracer::ntr = 11 The number of tracers in this module.
-
-
namespace
atmos_ocean_fluxes_mod¶ A dummy version of atmos_ocean_fluxes_mod module for use when the vastly larger FMS package is not needed.
Functions
-
integer function, public atmos_ocean_fluxes_mod::aof_set_coupler_flux(name name, flux_type flux_type, implementation implementation, atm_tr_index atm_tr_index, param param, flag flag, mol_wt mol_wt, ice_restart_file ice_restart_file, ocean_restart_file ocean_restart_file, units units, caller caller, verbosity verbosity) This subroutine duplicates an interface used by the FMS coupler, but only returns a value of -1. None of the arguments are used for anything.
- Parameters
[in] name: An unused argument[in] flux_type: An unused argument[in] implementation: An unused argument[in] atm_tr_index: An unused argument[in] param: An unused argument[in] flag: An unused argument[in] mol_wt: An unused argument[in] ice_restart_file: An unused argument[in] ocean_restart_file: An unused argument[in] units: An unused argument[in] caller: An unused argument[in] verbosity: An unused argument
-
-
namespace
baroclinic_zone_initialization¶ Initial conditions for an idealized baroclinic zone.
Functions
-
subroutine
bcz_params(G G, GV GV, US US, param_file param_file, S_ref S_ref, dSdz dSdz, delta_S delta_S, dSdx dSdx, T_ref T_ref, dTdz dTdz, delta_T delta_T, dTdx dTdx, L_zone L_zone, just_read_params just_read_params)¶ Reads the parameters unique to this module.
- Parameters
[in] g: Grid structure[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: Parameter file handle[out] s_ref: Reference salinity [ppt][out] dsdz: Salinity stratification [ppt Z-1 ~> ppt m-1][out] delta_s: Salinity difference across baroclinic zone [ppt][out] dsdx: Linear salinity gradient [ppt m-1][out] t_ref: Reference temperature [degC][out] dtdz: Temperature stratification [degC Z-1 ~> degC m-1][out] delta_t: Temperature difference across baroclinic zone [degC][out] dtdx: Linear temperature gradient [degC m-1][out] l_zone: Width of baroclinic zone [m][in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine, public baroclinic_zone_initialization::baroclinic_zone_init_temperature_salinity(T T, S S, h h, G G, GV GV, US US, param_file param_file, just_read_params just_read_params) Initialization of temperature and salinity with the baroclinic zone initial conditions.
- Parameters
[in] g: Grid structure[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[out] t: Potential temperature [degC][out] s: Salinity [ppt][in] h: The model thicknesses [H ~> m or kg m-2][in] param_file: Parameter file handle[in] just_read_params: If present and true, this call will only read parameters without changing T & S.
Variables
-
character(len=40) baroclinic_zone_initialization::mdl = "baroclinic_zone_initialization" This module’s name.
-
subroutine
-
namespace
benchmark_initialization¶ Initialization for the “bench mark” configuration.
Functions
-
subroutine, public benchmark_initialization::benchmark_initialize_topography(D D, G G, param_file param_file, max_depth max_depth, US US) This subroutine sets up the benchmark test case topography.
- Parameters
[in] g: The dynamic horizontal grid type[out] d: Ocean bottom depth in m or [Z ~> m] if US is present[in] param_file: Parameter file structure[in] max_depth: Maximum model depth in the units of D[in] us: A dimensional unit scaling type
-
subroutine, public benchmark_initialization::benchmark_initialize_thickness(h h, G G, GV GV, US US, param_file param_file, eqn_of_state eqn_of_state, P_ref P_ref, just_read_params just_read_params) Initializes layer thicknesses for the benchmark test case, by finding the depths of interfaces in a specified latitude-dependent temperature profile with an exponentially decaying thermocline on top of a linear stratification.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[out] h: The thickness that is being initialized [H ~> m or kg m-2].[in] param_file: A structure indicating the open file to parse for model parameter values.eqn_of_state: integer that selects the equation of state.[in] p_ref: The coordinate-density reference pressure [Pa].[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine, public benchmark_initialization::benchmark_init_temperature_salinity(T T, S S, G G, GV GV, param_file param_file, eqn_of_state eqn_of_state, P_Ref P_Ref, just_read_params just_read_params) Initializes layer temperatures and salinities for benchmark.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[out] t: The potential temperature that is being initialized.[out] s: The salinity that is being initialized.[in] param_file: A structure indicating the open file to parse for model parameter values.eqn_of_state: integer that selects the equation of state.[in] p_ref: The coordinate-density reference pressure [Pa].[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
-
namespace
bfb_initialization¶ Initialization of the boundary-forced-basing configuration.
Functions
-
subroutine, public bfb_initialization::bfb_set_coord(Rlay Rlay, g_prime g_prime, GV GV, param_file param_file, eqn_of_state eqn_of_state) This subroutine specifies the vertical coordinate in terms of temperature at the surface and at the bottom. This case is set up in such a way that the temperature of the topmost layer is equal to the SST at the southern edge of the domain. The temperatures are then converted to densities of the top and bottom layers and linearly interpolated for the intermediate layers.
- Parameters
[out] rlay: Layer potential density.[out] g_prime: The reduced gravity at each interface [L2 Z-1 T-2 ~> m s-2].[in] gv: The ocean’s vertical grid structure[in] param_file: A structure to parse for run-time parameterseqn_of_state: Integer that selects the equation of state.
-
subroutine, public bfb_initialization::bfb_initialize_sponges_southonly(G G, GV GV, US US, use_temperature use_temperature, tv tv, param_file param_file, CSp CSp, h h) This subroutine sets up the sponges for the southern bouundary of the domain. Maximum damping occurs within 2 degrees lat of the boundary. The damping linearly decreases northward over the next 2 degrees.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] use_temperature: If true, temperature and salinity are used as state variables.[in] tv: A structure pointing to various thermodynamic variables[in] param_file: A structure to parse for run-time parameterscsp: A pointer to the sponge control structure[in] h: Layer thicknesses [H ~> m or kg m-2]
-
subroutine
write_bfb_log(param_file param_file)¶ Write output about the parameter values being used.
- Parameters
[in] param_file: A structure indicating the open file to parse for model parameter values.
Variables
-
logical
first_call= .true.¶ Unsafe model variable.
-
-
namespace
bfb_surface_forcing¶ Surface forcing for the boundary-forced-basin (BFB) configuration.
Functions
-
subroutine, public bfb_surface_forcing::bfb_buoyancy_forcing(state state, fluxes fluxes, day day, dt dt, G G, US US, CS CS) Bouyancy forcing for the boundary-forced-basin (BFB) configuration.
- Parameters
[inout] state: A structure containing fields that describe the surface state of the ocean.[inout] fluxes: A structure containing pointers to any possible forcing fields. Unused fields have NULL ptrs.[in] day: Time of the fluxes.[in] dt: The amount of time over which the fluxes apply [s][in] g: The ocean’s grid structure[in] us: A dimensional unit scaling typecs: A pointer to the control structure returned by a previous call to BFB_surface_forcing_init.
-
subroutine, public bfb_surface_forcing::bfb_surface_forcing_init(Time Time, G G, US US, param_file param_file, diag diag, CS CS) Initialization for forcing the boundary-forced-basin (BFB) configuration.
- Parameters
[in] time: The current model time.[in] g: The ocean’s grid structure[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters[in] diag: A structure that is used to regulate diagnostic output.cs: A pointer to the control structure for this module
-
-
namespace
boundary_impulse_tracer¶ Implements a boundary impulse response tracer to calculate Green’s functions.
Functions
-
logical function, public boundary_impulse_tracer::register_boundary_impulse_tracer(HI HI, GV GV, param_file param_file, CS CS, tr_Reg tr_Reg, restart_CS restart_CS) Read in runtime options and add boundary impulse tracer to tracer registry.
- Parameters
[in] hi: A horizontal index type structure[in] gv: The ocean’s vertical grid structure[in] param_file: A structure to parse for run-time parameterscs: The control structure returned by a previous call to register_boundary_impulse_tracer.tr_reg: A pointer that is set to point to the control structure for the tracer advection and diffusion modulerestart_cs: A pointer to the restart control structure
-
subroutine, public boundary_impulse_tracer::initialize_boundary_impulse_tracer(restart restart, day day, G G, GV GV, h h, diag diag, OBC OBC, CS CS, sponge_CSp sponge_CSp, tv tv) Initialize tracer from restart or set to 1 at surface to initialize.
- Parameters
[in] restart: .true. if the fields have already been read from a restart file.[in] day: Time of the start of the run.[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][in] diag: A structure that is used to regulate diagnostic output.obc: This open boundary condition type specifies whether, where, and what open boundary conditions are used.cs: The control structure returned by a previous call to register_boundary_impulse_tracer.sponge_csp: Pointer to the control structure for the sponges.[in] tv: A structure pointing to various thermodynamic variables
-
subroutine, public boundary_impulse_tracer::boundary_impulse_tracer_column_physics(h_old h_old, h_new h_new, ea ea, eb eb, fluxes fluxes, dt dt, G G, GV GV, CS CS, tv tv, debug debug, evap_CFL_limit evap_CFL_limit, minimum_forcing_depth minimum_forcing_depth) Apply source or sink at boundary and do vertical diffusion.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h_old: Layer thickness before entrainment [H ~> m or kg m-2].[in] h_new: Layer thickness after entrainment [H ~> m or kg m-2].[in] ea: an array to which the amount of fluid entrained[in] eb: an array to which the amount of fluid entrained[in] fluxes: A structure containing pointers to thermodynamic and tracer forcing fields. Unused fields have NULL ptrs.[in] dt: The amount of time covered by this call [s]cs: The control structure returned by a previous call to register_boundary_impulse_tracer.[in] tv: A structure pointing to various thermodynamic variables[in] debug: If true calculate checksums[in] evap_cfl_limit: Limit on the fraction of the water that can be fluxed out of the top layer in a timestep [nondim][in] minimum_forcing_depth: The smallest depth over which fluxes can be applied [m]
-
integer function, public boundary_impulse_tracer::boundary_impulse_stock(h h, stocks stocks, G G, GV GV, CS CS, names names, units units, stock_index stock_index) Calculate total inventory of tracer.
- Return
Return value: the number of stocks calculated here.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][out] stocks: the mass-weighted integrated amount of each tracer, in kg times concentration units [kg conc].cs: The control structure returned by a previous call to register_boundary_impulse_tracer.[out] names: The names of the stocks calculated.[out] units: The units of the stocks calculated.[in] stock_index: The coded index of a specific stock being sought.
-
subroutine, public boundary_impulse_tracer::boundary_impulse_tracer_surface_state(state state, h h, G G, CS CS) This subroutine extracts the surface fields from this tracer package that are to be shared with the atmosphere in coupled configurations. This particular tracer package does not report anything back to the coupler.
- Parameters
[in] g: The ocean’s grid structure.[inout] state: A structure containing fields that describe the surface state of the ocean.[in] h: Layer thickness [H ~> m or kg m-2].cs: The control structure returned by a previous call to register_boundary_impulse_tracer.
-
subroutine, public boundary_impulse_tracer::boundary_impulse_tracer_end(CS CS) Performs finalization of boundary impulse tracer.
- Parameters
cs: The control structure returned by a previous call to register_boundary_impulse_tracer.
Variables
-
integer, parameter boundary_impulse_tracer::ntr_max = 1 NTR_MAX is the maximum number of tracers in this module.
-
-
namespace
circle_obcs_initialization¶ Configures the model for the “circle_obcs” experiment which tests Open Boundary Conditions radiating an SSH anomaly.
Functions
-
subroutine, public circle_obcs_initialization::circle_obcs_initialize_thickness(h h, G G, GV GV, param_file param_file, just_read_params just_read_params) This subroutine initializes layer thicknesses for the circle_obcs experiment.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[out] h: The thickness that is being initialized [H ~> m or kg m-2].[in] param_file: A structure indicating the open file to parse for model parameter values.[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
-
namespace
coord_adapt¶ Regrid columns for the adaptive coordinate.
Functions
-
subroutine, public coord_adapt::init_coord_adapt(CS CS, nk nk, coordinateResolution coordinateResolution, m_to_H m_to_H) Initialise an adapt_CS with parameters.
- Parameters
cs: Unassociated pointer to hold the control structure[in] nk: Number of layers in the grid[in] coordinateresolution: Nominal near-surface resolution [m] or other units specified with m_to_H[in] m_to_h: A conversion factor from m to the units of thicknesses
-
subroutine, public coord_adapt::end_coord_adapt(CS CS) Clean up the coordinate control structure.
- Parameters
cs: The control structure for this module
-
subroutine, public coord_adapt::set_adapt_params(CS CS, adaptTimeRatio adaptTimeRatio, adaptAlpha adaptAlpha, adaptZoom adaptZoom, adaptZoomCoeff adaptZoomCoeff, adaptBuoyCoeff adaptBuoyCoeff, adaptDrho0 adaptDrho0, adaptDoMin adaptDoMin) This subtroutine can be used to set the parameters for coord_adapt module.
- Parameters
cs: The control structure for this module[in] adapttimeratio: Ratio of optimisation and diffusion timescales[in] adaptalpha: Nondimensional coefficient determining how much optimisation to apply[in] adaptzoom: Near-surface zooming depth [H ~> m or kg m-2][in] adaptzoomcoeff: Near-surface zooming coefficient[in] adaptbuoycoeff: Stratification-dependent diffusion coefficient[in] adaptdrho0: Reference density difference for stratification-dependent diffusion[in] adaptdomin: If true, form a HYCOM1-like mixed layer by preventing interfaces from becoming shallower than the depths set by coordinateResolution
-
subroutine, public coord_adapt::build_adapt_column(CS CS, G G, GV GV, tv tv, i i, j j, zInt zInt, tInt tInt, sInt sInt, h h, zNext zNext) - Parameters
[in] cs: The control structure for this module[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] tv: A structure pointing to various thermodynamic variables[in] i: The i-index of the column to work on[in] j: The j-index of the column to work on[in] zint: Interface heights [H ~> m or kg m-2].[in] tint: Interface temperatures [degC][in] sint: Interface salinities [ppt][in] h: Layer thicknesses [H ~> m or kg m-2][inout] znext: updated interface positions
-
-
namespace
coord_hycom¶ Regrid columns for the HyCOM coordinate.
Functions
-
subroutine, public coord_hycom::init_coord_hycom(CS CS, nk nk, coordinateResolution coordinateResolution, target_density target_density, interp_CS interp_CS) Initialise a hycom_CS with pointers to parameters.
- Parameters
cs: Unassociated pointer to hold the control structure[in] nk: Number of layers in generated grid[in] coordinateresolution: Nominal near-surface resolution [m][in] target_density: Interface target densities [kg m-3][in] interp_cs: Controls for interpolation
-
subroutine, public coord_hycom::end_coord_hycom(CS CS) This subroutine deallocates memory in the control structure for the coord_hycom module.
- Parameters
cs: Coordinate control structure
-
subroutine, public coord_hycom::set_hycom_params(CS CS, max_interface_depths max_interface_depths, max_layer_thickness max_layer_thickness, interp_CS interp_CS) This subroutine can be used to set the parameters for the coord_hycom module.
- Parameters
cs: Coordinate control structure[in] max_interface_depths: Maximum depths of interfaces in m[in] max_layer_thickness: Maximum thicknesses of layers in m[in] interp_cs: Controls for interpolation
-
subroutine, public coord_hycom::build_hycom1_column(CS CS, eqn_of_state eqn_of_state, nz nz, depth depth, h h, T T, S S, p_col p_col, z_col z_col, z_col_new z_col_new, zScale zScale, h_neglect h_neglect, h_neglect_edge h_neglect_edge) Build a HyCOM coordinate column.
- Parameters
[in] cs: Coordinate control structureeqn_of_state: Equation of state structure[in] nz: Number of levels[in] depth: Depth of ocean bottom (positive [H ~> m or kg m-2])[in] t: Temperature of column [degC][in] s: Salinity of column [ppt][in] h: Layer thicknesses, in [m] or [H ~> m or kg m-2][in] p_col: Layer pressure [Pa][in] z_col: Interface positions relative to the surface [H ~> m or kg m-2][inout] z_col_new: Absolute positions of interfaces[in] zscale: Scaling factor from the input thicknesses in [m] to desired units for zInterface, perhaps m_to_H.[in] h_neglect: A negligibly small width for the purpose of cell reconstructions in the same units as h.[in] h_neglect_edge: A negligibly small width for the purpose of edge value calculations in the same units as h0.
-
-
namespace
coord_rho¶ Regrid columns for the continuous isopycnal (rho) coordinate.
Functions
-
subroutine, public coord_rho::init_coord_rho(CS CS, nk nk, ref_pressure ref_pressure, target_density target_density, interp_CS interp_CS) Initialise a rho_CS with pointers to parameters.
- Parameters
cs: Unassociated pointer to hold the control structure[in] nk: Number of layers in the grid[in] ref_pressure: Coordinate reference pressure [Pa][in] target_density: Nominal density of interfaces [kg m-3][in] interp_cs: Controls for interpolation
-
subroutine, public coord_rho::end_coord_rho(CS CS) This subroutine deallocates memory in the control structure for the coord_rho module.
- Parameters
cs: Coordinate control structure
-
subroutine, public coord_rho::set_rho_params(CS CS, min_thickness min_thickness, integrate_downward_for_e integrate_downward_for_e, interp_CS interp_CS) This subroutine can be used to set the parameters for the coord_rho module.
- Parameters
cs: Coordinate control structure[in] min_thickness: Minimum allowed thickness [H ~> m or kg m-2][in] integrate_downward_for_e: If true, integrate for interface positions from the top downward. If false, integrate from the bottom upward, as does the rest of the model.[in] interp_cs: Controls for interpolation
-
subroutine, public coord_rho::build_rho_column(CS CS, nz nz, depth depth, h h, T T, S S, eqn_of_state eqn_of_state, z_interface z_interface, h_neglect h_neglect, h_neglect_edge h_neglect_edge) Build a rho coordinate column.
Density profiles are calculated on the source grid.
Positions of target densities (for interfaces) are found by interpolation.
- Parameters
[in] cs: coord_rho control structure[in] nz: Number of levels on source grid (i.e. length of h, T, S)[in] depth: Depth of ocean bottom (positive in m)[in] h: Layer thicknesses [H ~> m or kg m-2][in] t: T for source column[in] s: S for source columneqn_of_state: Equation of state structure[inout] z_interface: Absolute positions of interfaces[in] h_neglect: A negligibly small width for the purpose of cell reconstructions in the same units as h0.[in] h_neglect_edge: A negligibly small width for the purpose of edge value calculations in the same units as h0.
-
subroutine
build_rho_column_iteratively(CS CS, remapCS remapCS, nz nz, depth depth, h h, T T, S S, eqn_of_state eqn_of_state, zInterface zInterface, h_neglect h_neglect, h_neglect_edge h_neglect_edge)¶ Iteratively build a rho coordinate column.
The algorithm operates as follows within each column:
Given T & S within each layer, the layer densities are computed.
Based on these layer densities, a global density profile is reconstructed (this profile is monotonically increasing and may be discontinuous)
The new grid interfaces are determined based on the target interface densities.
T & S are remapped onto the new grid.
Return to step 1 until convergence or until the maximum number of iterations is reached, whichever comes first.
- Parameters
[in] cs: Regridding control structure[in] remapcs: Remapping parameters and options[in] nz: Number of levels[in] depth: Depth of ocean bottom [Z ~> m][in] h: Layer thicknesses in Z coordinates [Z ~> m][in] t: T for column [degC][in] s: S for column [ppt]eqn_of_state: Equation of state structure[inout] zinterface: Absolute positions of interfaces[in] h_neglect: A negligibly small width for the purpose of cell reconstructions in the same units as h [Z ~> m][in] h_neglect_edge: A negligibly small width for the purpose of edge value calculations in the same units as h [Z ~> m]
-
subroutine
copy_finite_thicknesses(nk nk, h_in h_in, threshold threshold, nout nout, h_out h_out, mapping mapping)¶ Copy column thicknesses with vanished layers removed.
- Parameters
[in] nk: Number of layer for h_in, T_in, S_in[in] h_in: Thickness of input column[in] threshold: Thickness threshold defining vanished layers[out] nout: Number of non-vanished layers[out] h_out: Thickness of output column[out] mapping: Index of k-out corresponding to k-in
-
subroutine, public coord_rho::old_inflate_layers_1d(min_thickness min_thickness, nk nk, h h) Inflate vanished layers to finite (nonzero) width.
- Parameters
[in] min_thickness: Minimum allowed thickness [H ~> m or kg m-2][in] nk: Number of layers in the grid[inout] h: Layer thicknesses [H ~> m or kg m-2]
Variables
-
integer, parameter coord_rho::nb_regridding_iterations = 1 Maximum number of regridding iterations.
-
real, parameter coord_rho::deviation_tolerance = 1e-10 Deviation tolerance between succesive grids in regridding iterations.
-
-
namespace
coord_sigma¶ Regrid columns for the sigma coordinate.
Functions
-
subroutine, public coord_sigma::init_coord_sigma(CS CS, nk nk, coordinateResolution coordinateResolution) Initialise a sigma_CS with pointers to parameters.
- Parameters
cs: Unassociated pointer to hold the control structure[in] nk: Number of layers in the grid[in] coordinateresolution: Nominal coordinate resolution [nondim]
-
subroutine, public coord_sigma::end_coord_sigma(CS CS) This subroutine deallocates memory in the control structure for the coord_sigma module.
- Parameters
cs: Coordinate control structure
-
subroutine, public coord_sigma::set_sigma_params(CS CS, min_thickness min_thickness) This subroutine can be used to set the parameters for the coord_sigma module.
- Parameters
cs: Coordinate control structure[in] min_thickness: Minimum allowed thickness [H ~> m or kg m-2]
-
subroutine, public coord_sigma::build_sigma_column(CS CS, depth depth, totalThickness totalThickness, zInterface zInterface) Build a sigma coordinate column.
- Parameters
[in] cs: Coordinate control structure[in] depth: Depth of ocean bottom (positive [H ~> m or kg m-2])[in] totalthickness: Column thickness (positive [H ~> m or kg m-2])[inout] zinterface: Absolute positions of interfaces [H ~> m or kg m-2]
-
-
namespace
coord_slight¶ Regrid columns for the SLight coordinate.
Functions
-
subroutine, public coord_slight::init_coord_slight(CS CS, nk nk, ref_pressure ref_pressure, target_density target_density, interp_CS interp_CS, m_to_H m_to_H) Initialise a slight_CS with pointers to parameters.
- Parameters
cs: Unassociated pointer to hold the control structure[in] nk: Number of layers in the grid[in] ref_pressure: Coordinate reference pressure [Pa][in] target_density: Nominal density of interfaces [kg m-3][in] interp_cs: Controls for interpolation[in] m_to_h: A conversion factor from m to the units of thicknesses
-
subroutine, public coord_slight::end_coord_slight(CS CS) This subroutine deallocates memory in the control structure for the coord_slight module.
- Parameters
cs: Coordinate control structure
-
subroutine, public coord_slight::set_slight_params(CS CS, max_interface_depths max_interface_depths, max_layer_thickness max_layer_thickness, min_thickness min_thickness, compressibility_fraction compressibility_fraction, dz_ml_min dz_ml_min, nz_fixed_surface nz_fixed_surface, Rho_ML_avg_depth Rho_ML_avg_depth, nlay_ML_offset nlay_ML_offset, fix_haloclines fix_haloclines, halocline_filter_length halocline_filter_length, halocline_strat_tol halocline_strat_tol, interp_CS interp_CS) This subroutine can be used to set the parameters for the coord_slight module.
- Parameters
cs: Coordinate control structure[in] max_interface_depths: Maximum depths of interfaces [H ~> m or kg m-2][in] max_layer_thickness: Maximum thicknesses of layers [H ~> m or kg m-2][in] min_thickness: Minimum thickness allowed when building the new grid through regridding [H ~> m or kg m-2][in] compressibility_fraction: Fraction (between 0 and 1) of compressibility to add to potential density profiles when interpolating for target grid positions. [nondim][in] dz_ml_min: The fixed resolution in the topmost SLight_nkml_min layers [H ~> m or kg m-2][in] nz_fixed_surface: The number of fixed-thickness layers at the top of the model[in] rho_ml_avg_depth: Depth over which to average to determine the mixed layer potential density [H ~> m or kg m-2][in] nlay_ml_offset: Number of layers to offset the mixed layer density to find resolved stratification [nondim][in] fix_haloclines: If true, detect regions with much weaker than based on in-situ density, and use a stretched coordinate there.[in] halocline_filter_length: A length scale over which to filter T & S when looking for spuriously unstable water mass profiles [H ~> m or kg m-2].[in] halocline_strat_tol: A value of the stratification ratio that defines a problematic halocline region [nondim].[in] interp_cs: Controls for interpolation
-
subroutine, public coord_slight::build_slight_column(CS CS, eqn_of_state eqn_of_state, H_to_Pa H_to_Pa, H_subroundoff H_subroundoff, nz nz, depth depth, h_col h_col, T_col T_col, S_col S_col, p_col p_col, z_col z_col, z_col_new z_col_new, h_neglect h_neglect, h_neglect_edge h_neglect_edge) Build a SLight coordinate column.
- Parameters
[in] cs: Coordinate control structureeqn_of_state: Equation of state structure[in] h_to_pa: GVH_to_Pa[in] h_subroundoff: GVH_subroundoff[in] nz: Number of levels[in] depth: Depth of ocean bottom (positive [H ~> m or kg m-2])[in] t_col: T for column[in] s_col: S for column[in] h_col: Layer thicknesses [H ~> m or kg m-2][in] p_col: Layer quantities[in] z_col: Interface positions relative to the surface [H ~> m or kg m-2][inout] z_col_new: Absolute positions of interfaces [H ~> m or kg m-2][in] h_neglect: A negligibly small width for the purpose of cell reconstructions [H ~> m or kg m-2].[in] h_neglect_edge: A negligibly small width for the purpose of edge value calculations [H ~> m or kg m-2].
-
subroutine
rho_interfaces_col(rho_col rho_col, h_col h_col, z_col z_col, rho_tgt rho_tgt, nz nz, z_col_new z_col_new, CS CS, reliable reliable, debug debug, h_neglect h_neglect, h_neglect_edge h_neglect_edge)¶ Finds the new interface locations in a column of water that match the prescribed target densities.
- Parameters
[in] nz: Number of layers[in] rho_col: Initial layer reference densities.[in] h_col: Initial layer thicknesses.[in] z_col: Initial interface heights.[in] rho_tgt: Interface target densities.[inout] z_col_new: New interface heights.[in] cs: Coordinate control structure[inout] reliable: If true, the interface positions are well defined from a stable region.[in] debug: If present and true, do debugging checks.[in] h_neglect: A negligibly small width for the purpose of cell reconstructions in the same units as h_col.[in] h_neglect_edge: A negligibly small width for the purpose of edge value calculations in the same units as h_col.
-
-
namespace
coord_zlike¶ Regrid columns for a z-like coordinate (z-star, z-level)
Functions
-
subroutine, public coord_zlike::init_coord_zlike(CS CS, nk nk, coordinateResolution coordinateResolution) Initialise a zlike_CS with pointers to parameters.
- Parameters
cs: Unassociated pointer to hold the control structure[in] nk: Number of levels in the grid[in] coordinateresolution: Target coordinate resolution [Z ~> m]
-
subroutine, public coord_zlike::end_coord_zlike(CS CS) Deallocates the zlike control structure.
- Parameters
cs: Coordinate control structure
-
subroutine, public coord_zlike::set_zlike_params(CS CS, min_thickness min_thickness) Set parameters in the zlike structure.
- Parameters
cs: Coordinate control structure[in] min_thickness: Minimum allowed thickness [H ~> m or kg m-2]
-
subroutine, public coord_zlike::build_zstar_column(CS CS, depth depth, total_thickness total_thickness, zInterface zInterface, z_rigid_top z_rigid_top, eta_orig eta_orig, zScale zScale) Builds a z* coordinate with a minimum thickness.
- Parameters
[in] cs: Coordinate control structure[in] depth: Depth of ocean bottom (positive in the output units)[in] total_thickness: Column thickness (positive in the same units as depth)[inout] zinterface: Absolute positions of interfaces[in] z_rigid_top: The height of a rigid top (negative in the same units as depth)[in] eta_orig: The actual original height of the top in the same units as depth[in] zscale: Scaling factor from the target coordinate resolution in Z to desired units for zInterface, perhaps Z_to_H
-
-
namespace
dense_water_initialization¶ Initialization routines for the dense water formation and overflow experiment.
This experiment consists of a shelf accumulating dense water, which spills over an upward slope and a sill, before flowing down a slope into an open ocean region. It’s intended as a test of one of the motivating situations for the adaptive coordinate.
The nondimensional widths of the 5 regions are controlled by the
DENSE_WATER_DOMAIN_PARAMS, and the heights of the sill and shelf as a fraction of the total domain depth are controlled byDENSE_WATER_SILL_HEIGHTandDENSE_WATER_SHELF_HEIGHT.The density in the domain is governed by a linear equation of state, and is set up with a mixed layer of non-dimensional depth
DENSE_WATER_MLDbelow which there is a linear stratification fromS_REF, increasing byS_RANGEto the bottom.To force the experiment, there are sponges on the inflow and outflow of the domain. The inflow sponge has a salinity anomaly of
DENSE_WATER_EAST_SPONGE_SALTthrough the entire depth. The outflow sponge simply restores to the initial condition. Both sponges have controllable widths and restoring timescales.Functions
-
subroutine, public dense_water_initialization::dense_water_initialize_topography(D D, G G, param_file param_file, max_depth max_depth) Initialize the topography field for the dense water experiment.
- Parameters
[in] g: The dynamic horizontal grid type[out] d: Ocean bottom depth in the units of depth_max[in] param_file: Parameter file structure[in] max_depth: Maximum ocean depth in arbitrary units
-
subroutine, public dense_water_initialization::dense_water_initialize_ts(G G, GV GV, param_file param_file, eqn_of_state eqn_of_state, T T, S S, h h, just_read_params just_read_params) Initialize the temperature and salinity for the dense water experiment.
- Parameters
[in] g: Horizontal grid control structure[in] gv: Vertical grid control structure[in] param_file: Parameter file structureeqn_of_state: EOS structure[out] t: Output temperature [degC][out] s: Output salinity [ppt][in] h: Layer thicknesses [H ~> m or kg m-2][in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine, public dense_water_initialization::dense_water_initialize_sponges(G G, GV GV, tv tv, param_file param_file, use_ALE use_ALE, CSp CSp, ACSp ACSp) Initialize the restoring sponges for the dense water experiment.
- Parameters
[in] g: Horizontal grid control structure[in] gv: Vertical grid control structure[in] tv: Thermodynamic variables[in] param_file: Parameter file structure[in] use_ale: ALE flagcsp: Layered sponge control structure pointeracsp: ALE sponge control structure pointer
Variables
-
character(len=40) dense_water_initialization::mdl = "dense_water_initialization" Module name.
-
real, parameter dense_water_initialization::default_sill = 0.2 Default depth of the sill [nondim].
-
real, parameter dense_water_initialization::default_shelf = 0.4 Default depth of the shelf [nondim].
-
real, parameter dense_water_initialization::default_mld = 0.25 Default depth of the mixed layer [nondim].
-
-
namespace
dome2d_initialization¶ Initialization of the 2D DOME experiment with density water initialized on a coastal shelf.
Functions
-
subroutine, public dome2d_initialization::dome2d_initialize_topography(D D, G G, param_file param_file, max_depth max_depth) Initialize topography with a shelf and slope in a 2D domain.
- Parameters
[in] g: The dynamic horizontal grid type[out] d: Ocean bottom depth in the units of depth_max[in] param_file: Parameter file structure[in] max_depth: Maximum ocean depth in arbitrary units
-
subroutine, public dome2d_initialization::dome2d_initialize_thickness(h h, G G, GV GV, US US, param_file param_file, just_read_params just_read_params) Initialize thicknesses according to coordinate mode.
- Parameters
[in] g: Ocean grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[out] h: The thickness that is being initialized [H ~> m or kg m-2].[in] param_file: A structure indicating the open file to parse for model parameter values.[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine, public dome2d_initialization::dome2d_initialize_temperature_salinity(T T, S S, h h, G G, GV GV, param_file param_file, eqn_of_state eqn_of_state, just_read_params just_read_params) Initialize temperature and salinity in the 2d DOME configuration.
- Parameters
[in] g: Ocean grid structure[in] gv: The ocean’s vertical grid structure.[out] t: Potential temperature [degC][out] s: Salinity [ppt][in] h: Layer thickness [H ~> m or kg m-2][in] param_file: Parameter file structureeqn_of_state: Equation of state structure[in] just_read_params: If present and true, this call will only read parameters without changing T & S.
-
subroutine, public dome2d_initialization::dome2d_initialize_sponges(G G, GV GV, tv tv, param_file param_file, use_ALE use_ALE, CSp CSp, ACSp ACSp) Set up sponges in 2d DOME configuration.
- Parameters
[in] g: Ocean grid structure[in] gv: Vertical grid structure[in] tv: Thermodynamics structure[in] param_file: Parameter file structure[in] use_ale: If true, indicates model is in ALE modecsp: Layer-mode sponge structureacsp: ALE-mode sponge structure
Variables
-
character(len=40) dome2d_initialization::mdl = "DOME2D_initialization" This module’s name.
-
-
namespace
dome_initialization¶ Configures the model for the “DOME” experiment. DOME = Dynamics of Overflows and Mixing Experiment.
Functions
-
subroutine, public dome_initialization::dome_initialize_topography(D D, G G, param_file param_file, max_depth max_depth, US US) This subroutine sets up the DOME topography.
- Parameters
[in] g: The dynamic horizontal grid type[out] d: Ocean bottom depth in m or Z if US is present[in] param_file: Parameter file structure[in] max_depth: Maximum model depth in the units of D[in] us: A dimensional unit scaling type
-
subroutine, public dome_initialization::dome_initialize_thickness(h h, G G, GV GV, param_file param_file, just_read_params just_read_params) This subroutine initializes layer thicknesses for the DOME experiment.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[out] h: The thickness that is being initialized [H ~> m or kg m-2].[in] param_file: A structure indicating the open file to parse for model parameter values.[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine, public dome_initialization::dome_initialize_sponges(G G, GV GV, US US, tv tv, PF PF, CSp CSp) This subroutine sets the inverse restoration time (Idamp), and ! the values towards which the interface heights and an arbitrary ! number of tracers should be restored within each sponge. The ! interface height is always subject to damping, and must always be ! the first registered field. !
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] tv: A structure containing pointers to any available thermodynamic fields, including potential temperature and salinity or mixed layer density. Absent fields have NULL ptrs.[in] pf: A structure indicating the open file to parse for model parameter values.csp: A pointer that is set to point to the control structure for this module.
-
subroutine, public dome_initialization::dome_set_obc_data(OBC OBC, tv tv, G G, GV GV, US US, param_file param_file, tr_Reg tr_Reg) This subroutine sets the properties of flow at open boundary conditions. This particular example is for the DOME inflow describe in Legg et al. 2006.
- Parameters
obc: This open boundary condition type specifies whether, where, and what open boundary conditions are used.[in] tv: A structure containing pointers to any available thermodynamic fields, including potential temperature and salinity or mixed layer density. Absent fields have NULL ptrs.[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: A structure indicating the open file to parse for model parameter values.tr_reg: Tracer registry.
-
-
namespace
dome_tracer¶ A tracer package that is used as a diagnostic in the DOME experiments.
By Robert Hallberg, 2002
This file contains an example of the code that is needed to set up and use a set (in this case eleven) of dynamically passive tracers. These tracers dye the inflowing water or water initially within a range of latitudes or water initially in a range of depths.
A single subroutine is called from within each file to register each of the tracers for reinitialization and advection and to register the subroutine that initializes the tracers and set up their output and the subroutine that does any tracer physics or chemistry along with diapycnal mixing (included here because some tracers may float or swim vertically or dye diapycnal processes).
Functions
-
logical function, public dome_tracer::register_dome_tracer(HI HI, GV GV, param_file param_file, CS CS, tr_Reg tr_Reg, restart_CS restart_CS) Register tracer fields and subroutines to be used with MOM.
- Parameters
[in] hi: A horizontal index type structure.[in] gv: The ocean’s vertical grid structure[in] param_file: A structure to parse for run-time parameterscs: A pointer that is set to point to the control structure for this moduletr_reg: A pointer to the tracer registry.restart_cs: A pointer to the restart control structure.
-
subroutine, public dome_tracer::initialize_dome_tracer(restart restart, day day, G G, GV GV, US US, h h, diag diag, OBC OBC, CS CS, sponge_CSp sponge_CSp, param_file param_file) Initializes the NTR tracer fields in tr(:,:,:,:) and sets up the tracer output.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[in] restart: .true. if the fields have already been read from a restart file.[in] day: Time of the start of the run.[in] h: Layer thicknesses [H ~> m or kg m-2][in] diag: Structure used to regulate diagnostic output.obc: Structure specifying open boundary options.cs: The control structure returned by a previous call to DOME_register_tracer.sponge_csp: A pointer to the control structure for the sponges, if they are in use.[in] param_file: A structure to parse for run-time parameters
-
subroutine, public dome_tracer::dome_tracer_column_physics(h_old h_old, h_new h_new, ea ea, eb eb, fluxes fluxes, dt dt, G G, GV GV, CS CS, evap_CFL_limit evap_CFL_limit, minimum_forcing_depth minimum_forcing_depth) This subroutine applies diapycnal diffusion and any other column tracer physics or chemistry to the tracers from this file. This is a simple example of a set of advected passive tracers.
The arguments to this subroutine are redundant in that h_new(k) = h_old(k) + ea(k) - eb(k-1) + eb(k) - ea(k+1)
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h_old: Layer thickness before entrainment [H ~> m or kg m-2].[in] h_new: Layer thickness after entrainment [H ~> m or kg m-2].[in] ea: an array to which the amount of fluid entrained[in] eb: an array to which the amount of fluid entrained[in] fluxes: A structure containing pointers to thermodynamic and tracer forcing fields. Unused fields have NULL ptrs.[in] dt: The amount of time covered by this call [s]cs: The control structure returned by a previous call to DOME_register_tracer.[in] evap_cfl_limit: Limit on the fraction of the water that can be fluxed out of the top layer in a timestep [nondim][in] minimum_forcing_depth: The smallest depth over which fluxes can be applied [m]
-
subroutine, public dome_tracer::dome_tracer_surface_state(state state, h h, G G, CS CS) This subroutine extracts the surface fields from this tracer package that are to be shared with the atmosphere in coupled configurations. This particular tracer package does not report anything back to the coupler.
- Parameters
[in] g: The ocean’s grid structure.[inout] state: A structure containing fields that describe the surface state of the ocean.[in] h: Layer thickness [H ~> m or kg m-2].cs: The control structure returned by a previous call to DOME_register_tracer.
-
subroutine, public dome_tracer::dome_tracer_end(CS CS) Clean up memory allocations, if any.
- Parameters
cs: The control structure returned by a previous call to DOME_register_tracer.
Variables
-
integer, parameter dome_tracer::ntr = 11 The number of tracers in this module.
-
-
namespace
dumbbell_initialization¶ Configures the model for the idealized dumbbell test case.
Functions
-
subroutine, public dumbbell_initialization::dumbbell_initialize_topography(D D, G G, param_file param_file, max_depth max_depth) Initialization of topography.
- Parameters
[in] g: The dynamic horizontal grid type[out] d: Ocean bottom depth in the units of depth_max[in] param_file: Parameter file structure[in] max_depth: Maximum ocean depth in arbitrary units
-
subroutine, public dumbbell_initialization::dumbbell_initialize_thickness(h h, G G, GV GV, US US, param_file param_file, just_read_params just_read_params) Initializes the layer thicknesses to be uniform in the dumbbell test case.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[out] h: The thickness that is being initialized [H ~> m or kg m-2].[in] param_file: A structure indicating the open file to parse for model parameter values.[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine, public dumbbell_initialization::dumbbell_initialize_temperature_salinity(T T, S S, h h, G G, GV GV, param_file param_file, eqn_of_state eqn_of_state, just_read_params just_read_params) Initial values for temperature and salinity for the dumbbell test case.
- Parameters
[in] g: Ocean grid structure[in] gv: Vertical grid structure[out] t: Potential temperature [degC][out] s: Salinity [ppt][in] h: Layer thickness [H ~> m or kg m-2][in] param_file: Parameter file structureeqn_of_state: Equation of state structure[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine, public dumbbell_initialization::dumbbell_initialize_sponges(G G, GV GV, US US, tv tv, param_file param_file, use_ALE use_ALE, CSp CSp, ACSp ACSp) Initialize the restoring sponges for the dumbbell test case.
- Parameters
[in] g: Horizontal grid control structure[in] gv: Vertical grid control structure[in] us: A dimensional unit scaling type[in] tv: Thermodynamic variables[in] param_file: Parameter file structure[in] use_ale: ALE flagcsp: Layered sponge control structure pointeracsp: ALE sponge control structure pointer
Variables
-
character(len=40) dumbbell_initialization::mdl = "dumbbell_initialization" This module’s name.
-
-
namespace
dumbbell_surface_forcing¶ Surface forcing for the dumbbell test case.
Functions
-
subroutine, public dumbbell_surface_forcing::dumbbell_buoyancy_forcing(state state, fluxes fluxes, day day, dt dt, G G, CS CS) Surface buoyancy (heat and fresh water) fluxes for the dumbbell test case.
- Parameters
[inout] state: A structure containing fields that describe the surface state of the ocean.[inout] fluxes: A structure containing pointers to any possible forcing fields. Unused fields have NULL ptrs.[in] day: Time of the fluxes.[in] dt: The amount of time over which the fluxes apply [s][in] g: The ocean’s grid structurecs: A control structure returned by a previous call to dumbbell_surface_forcing_init
-
subroutine, public dumbbell_surface_forcing::dumbbell_dynamic_forcing(state state, fluxes fluxes, day day, dt dt, G G, CS CS) Dynamic forcing for the dumbbell test case.
- Parameters
[inout] state: A structure containing fields that describe the surface state of the ocean.[inout] fluxes: A structure containing pointers to any possible forcing fields. Unused fields have NULL ptrs.[in] day: Time of the fluxes.[in] dt: The amount of time over which the fluxes apply [s][in] g: The ocean’s grid structurecs: A control structure returned by a previous call to dumbbell_surface_forcing_init
-
subroutine, public dumbbell_surface_forcing::dumbbell_surface_forcing_init(Time Time, G G, US US, param_file param_file, diag diag, CS CS) Reads and sets up the forcing for the dumbbell test case.
- Parameters
[in] time: The current model time.[in] g: The ocean’s grid structure[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters[in] diag: A structure that is used to regulate diagnostic output.cs: A pointer to the control structure for this module
-
-
namespace
dyed_channel_initialization¶ Initialization for the dyed_channel configuration.
Setting dyes, one for painting the inflow on each side.
Functions
-
logical function, public dyed_channel_initialization::register_dyed_channel_obc(param_file param_file, CS CS, OBC_Reg OBC_Reg) Add dyed channel to OBC registry.
- Parameters
[in] param_file: parameter file.cs: Dyed channel control structure.obc_reg: OBC registry.
-
subroutine, public dyed_channel_initialization::dyed_channel_obc_end(CS CS) Clean up the dyed_channel OBC from registry.
- Parameters
cs: Dyed channel control structure.
-
subroutine, public dyed_channel_initialization::dyed_channel_set_obc_tracer_data(OBC OBC, G G, GV GV, param_file param_file, tr_Reg tr_Reg) This subroutine sets the dye and flow properties at open boundary conditions.
- Parameters
obc: This open boundary condition type specifies whether, where, and what open boundary conditions are used.[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] param_file: A structure indicating the open file to parse for model parameter values.tr_reg: Tracer registry.
-
subroutine, public dyed_channel_initialization::dyed_channel_update_flow(OBC OBC, CS CS, G G, Time Time) This subroutine updates the long-channel flow.
- Parameters
obc: This open boundary condition type specifies whether, where, and what open boundary conditions are used.cs: Dyed channel control structure.[in] g: The ocean’s grid structure.[in] time: model time.
Variables
-
integer
ntr= 0¶ Number of dye tracers.
-
-
namespace
dyed_obc_tracer¶ This tracer package dyes flow through open boundaries.
By Kate Hedstrom, 2017, copied from DOME tracers and also dye_example.
This file contains an example of the code that is needed to set up and use a set of dynamically passive tracers. These tracers dye the inflowing water, one per open boundary segment.
A single subroutine is called from within each file to register each of the tracers for reinitialization and advection and to register the subroutine that initializes the tracers and set up their output and the subroutine that does any tracer physics or chemistry along with diapycnal mixing (included here because some tracers may float or swim vertically or dye diapycnal processes).
Functions
-
logical function, public dyed_obc_tracer::register_dyed_obc_tracer(HI HI, GV GV, param_file param_file, CS CS, tr_Reg tr_Reg, restart_CS restart_CS) Register tracer fields and subroutines to be used with MOM.
- Parameters
[in] hi: A horizontal index type structure.[in] gv: The ocean’s vertical grid structure[in] param_file: A structure to parse for run-time parameterscs: A pointer that is set to point to the control structure for this moduletr_reg: A pointer to the tracer registry.restart_cs: A pointer to the restart control structure.
-
subroutine, public dyed_obc_tracer::initialize_dyed_obc_tracer(restart restart, day day, G G, GV GV, h h, diag diag, OBC OBC, CS CS) Initializes the CSntr tracer fields in tr(:,:,:,:) and sets up the tracer output.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] restart: .true. if the fields have already been read from a restart file.[in] day: Time of the start of the run.[in] h: Layer thicknesses [H ~> m or kg m-2][in] diag: Structure used to regulate diagnostic output.obc: Structure specifying open boundary options.cs: The control structure returned by a previous call to dyed_obc_register_tracer.
-
subroutine, public dyed_obc_tracer::dyed_obc_tracer_column_physics(h_old h_old, h_new h_new, ea ea, eb eb, fluxes fluxes, dt dt, G G, GV GV, CS CS, evap_CFL_limit evap_CFL_limit, minimum_forcing_depth minimum_forcing_depth) This subroutine applies diapycnal diffusion and any other column tracer physics or chemistry to the tracers from this file. This is a simple example of a set of advected passive tracers.
The arguments to this subroutine are redundant in that h_new(k) = h_old(k) + ea(k) - eb(k-1) + eb(k) - ea(k+1)
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h_old: Layer thickness before entrainment [H ~> m or kg m-2].[in] h_new: Layer thickness after entrainment [H ~> m or kg m-2].[in] ea: an array to which the amount of fluid entrained[in] eb: an array to which the amount of fluid entrained[in] fluxes: A structure containing pointers to thermodynamic and tracer forcing fields. Unused fields have NULL ptrs.[in] dt: The amount of time covered by this call [s]cs: The control structure returned by a previous call to dyed_obc_register_tracer.[in] evap_cfl_limit: Limit on the fraction of the water that can be fluxed out of the top layer in a timestep [nondim][in] minimum_forcing_depth: The smallest depth over which fluxes can be applied [m]
-
subroutine, public dyed_obc_tracer::dyed_obc_tracer_end(CS CS) Clean up memory allocations, if any.
- Parameters
cs: The control structure returned by a previous call to dyed_obc_register_tracer.
-
-
namespace
dyed_obcs_initialization¶ Dyed open boundary conditions.
Setting dyes, one for painting the inflow on each side.
Functions
-
subroutine, public dyed_obcs_initialization::dyed_obcs_set_obc_data(OBC OBC, G G, GV GV, param_file param_file, tr_Reg tr_Reg) This subroutine sets the dye properties at open boundary conditions.
- Parameters
obc: This open boundary condition type specifies whether, where, and what open boundary conditions are used.[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] param_file: A structure indicating the open file to parse for model parameter values.tr_reg: Tracer registry.
Variables
-
integer
ntr= 0¶ Number of dye tracers.
-
-
namespace
external_gwave_initialization¶ Initialization for the “external gravity wave wave” configuration.
Functions
-
subroutine, public external_gwave_initialization::external_gwave_initialize_thickness(h h, G G, GV GV, US US, param_file param_file, just_read_params just_read_params) This subroutine initializes layer thicknesses for the external_gwave experiment.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[out] h: The thickness that is being initialized [H ~> m or kg m-2].[in] param_file: A structure indicating the open file to parse for model parameter values.[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
-
namespace
ideal_age_example¶ A tracer package of ideal age tracers.
Originally by Robert Hallberg, 2002
This file contains an example of the code that is needed to set up and use a set (in this case two) of dynamically passive tracers for diagnostic purposes. The tracers here are an ideal age tracer that ages at a rate of 1/year once it is isolated from the surface, and a vintage tracer, whose surface concentration grows exponen- with time with a 30-year timescale (similar to CFCs).
Functions
-
logical function, public ideal_age_example::register_ideal_age_tracer(HI HI, GV GV, param_file param_file, CS CS, tr_Reg tr_Reg, restart_CS restart_CS) Register the ideal age tracer fields to be used with MOM.
- Parameters
[in] hi: A horizontal index type structure[in] gv: The ocean’s vertical grid structure[in] param_file: A structure to parse for run-time parameterscs: The control structure returned by a previous call to register_ideal_age_tracer.tr_reg: A pointer that is set to point to the control structure for the tracer advection and diffusion modulerestart_cs: A pointer to the restart control structure
-
subroutine, public ideal_age_example::initialize_ideal_age_tracer(restart restart, day day, G G, GV GV, US US, h h, diag diag, OBC OBC, CS CS, sponge_CSp sponge_CSp) Sets the ideal age traces to their initial values and sets up the tracer output.
- Parameters
[in] restart: .true. if the fields have already been read from a restart file.[in] day: Time of the start of the run.[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[in] h: Layer thicknesses [H ~> m or kg m-2][in] diag: A structure that is used to regulate diagnostic output.obc: This open boundary condition type specifies whether, where, and what open boundary conditions are used.cs: The control structure returned by a previous call to register_ideal_age_tracer.sponge_csp: Pointer to the control structure for the sponges.
-
subroutine, public ideal_age_example::ideal_age_tracer_column_physics(h_old h_old, h_new h_new, ea ea, eb eb, fluxes fluxes, dt dt, G G, GV GV, CS CS, evap_CFL_limit evap_CFL_limit, minimum_forcing_depth minimum_forcing_depth) Applies diapycnal diffusion, aging and regeneration at the surface to the ideal age tracers.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h_old: Layer thickness before entrainment [H ~> m or kg m-2].[in] h_new: Layer thickness after entrainment [H ~> m or kg m-2].[in] ea: an array to which the amount of fluid entrained[in] eb: an array to which the amount of fluid entrained[in] fluxes: A structure containing pointers to thermodynamic and tracer forcing fields. Unused fields have NULL ptrs.[in] dt: The amount of time covered by this call [s]cs: The control structure returned by a previous call to register_ideal_age_tracer.[in] evap_cfl_limit: Limit on the fraction of the water that can be fluxed out of the top layer in a timestep [nondim][in] minimum_forcing_depth: The smallest depth over which fluxes can be applied [m]
-
integer function, public ideal_age_example::ideal_age_stock(h h, stocks stocks, G G, GV GV, CS CS, names names, units units, stock_index stock_index) Calculates the mass-weighted integral of all tracer stocks, returning the number of stocks it has calculated. If stock_index is present, only the stock corresponding to that coded index is found.
- Return
The number of stocks calculated here.
- Parameters
[in] g: The ocean’s grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][out] stocks: the mass-weighted integrated amount of each tracer, in kg times concentration units [kg conc].[in] gv: The ocean’s vertical grid structurecs: The control structure returned by a previous call to register_ideal_age_tracer.[out] names: the names of the stocks calculated.[out] units: the units of the stocks calculated.[in] stock_index: the coded index of a specific stock being sought.
-
subroutine, public ideal_age_example::ideal_age_tracer_surface_state(state state, h h, G G, CS CS) This subroutine extracts the surface fields from this tracer package that are to be shared with the atmosphere in coupled configurations. This particular tracer package does not report anything back to the coupler.
- Parameters
[in] g: The ocean’s grid structure.[inout] state: A structure containing fields that describe the surface state of the ocean.[in] h: Layer thickness [H ~> m or kg m-2].cs: The control structure returned by a previous call to register_ideal_age_tracer.
-
subroutine, public ideal_age_example::ideal_age_example_end(CS CS) Deallocate any memory associated with this tracer package.
- Parameters
cs: The control structure returned by a previous call to register_ideal_age_tracer.
Variables
-
integer, parameter ideal_age_example::ntr_max = 3 the maximum number of tracers in this module.
-
-
namespace
idealized_hurricane¶ Forcing for the idealized hurricane and SCM_idealized_hurricane examples.
Functions
-
subroutine, public idealized_hurricane::idealized_hurricane_wind_init(Time Time, G G, param_file param_file, CS CS) Initializes wind profile for the SCM idealized hurricane example.
- Parameters
[in] time: Model time[in] g: Grid structure[in] param_file: Input parameter structurecs: Parameter container
-
subroutine, public idealized_hurricane::idealized_hurricane_wind_forcing(state state, forces forces, day day, G G, US US, CS CS) Computes the surface wind for the idealized hurricane test cases.
- Parameters
[in] state: Surface state structure[inout] forces: A structure with the driving mechanical forces[in] day: Time in days[inout] g: Grid structure[in] us: A dimensional unit scaling typecs: Container for idealized hurricane parameters
-
subroutine
idealized_hurricane_wind_profile(CS CS, absf absf, YY YY, XX XX, UOCN UOCN, VOCN VOCN, Tx Tx, Ty Ty)¶ Calculate the wind speed at a location as a function of time.
- Parameters
cs: Container for SCM parameters[in] absf: Input coriolis magnitude[in] yy: Location in m relative to center y[in] xx: Location in m relative to center x[in] uocn: X surface current[in] vocn: Y surface current[out] tx: X stress[out] ty: Y stress
-
subroutine, public idealized_hurricane::scm_idealized_hurricane_wind_forcing(state state, forces forces, day day, G G, US US, CS CS) This subroutine is primarily needed as a legacy for reproducing answers. It is included as an additional subroutine rather than padded into the previous routine with flags to ease its eventual removal. Its functionality is replaced with the new routines and it can be deleted when answer changes are acceptable.
- Parameters
[in] state: Surface state structure[inout] forces: A structure with the driving mechanical forces[in] day: Time in days[inout] g: Grid structure[in] us: A dimensional unit scaling typecs: Container for SCM parameters
Variables
-
character(len=40) idealized_hurricane::mdl = "idealized_hurricane" This module’s name.
-
-
namespace
isomip_initialization¶ Configures the ISOMIP test case.
See this paper for details: http://www.geosci-model-dev-discuss.net/8/9859/2015/gmdd-8-9859-2015.pdf
Functions
-
subroutine, public isomip_initialization::isomip_initialize_topography(D D, G G, param_file param_file, max_depth max_depth, US US) Initialization of topography for the ISOMIP configuration.
- Parameters
[in] g: The dynamic horizontal grid type[out] d: Ocean bottom depth in m or Z if US is present[in] param_file: Parameter file structure[in] max_depth: Maximum model depth in the units of D[in] us: A dimensional unit scaling type
-
subroutine, public isomip_initialization::isomip_initialize_thickness(h h, G G, GV GV, US US, param_file param_file, tv tv, just_read_params just_read_params) Initialization of thicknesses.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[out] h: The thickness that is being initialized [H ~> m or kg m-2].[in] param_file: A structure indicating the open file to parse for model parameter values.[in] tv: A structure containing pointers to any available thermodynamic fields, including the eqn. of state.[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine, public isomip_initialization::isomip_initialize_temperature_salinity(T T, S S, h h, G G, GV GV, param_file param_file, eqn_of_state eqn_of_state, just_read_params just_read_params) Initial values for temperature and salinity.
- Parameters
[in] g: Ocean grid structure[in] gv: Vertical grid structure[out] t: Potential temperature [degC][out] s: Salinity [ppt][in] h: Layer thickness [H ~> m or kg m-2][in] param_file: Parameter file structureeqn_of_state: Equation of state structure[in] just_read_params: If present and true, this call will only read parameters without changing T & S.
-
subroutine, public isomip_initialization::isomip_initialize_sponges(G G, GV GV, US US, tv tv, PF PF, use_ALE use_ALE, CSp CSp, ACSp ACSp) Sets up the the inverse restoration time (Idamp), and.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] tv: A structure containing pointers to any available thermodynamic fields, potential temperature and salinity or mixed layer density. Absent fields have NULL ptrs.[in] pf: A structure indicating the open file to parse for model parameter values.[in] use_ale: If true, indicates model is in ALE modecsp: Layer-mode sponge structureacsp: ALE-mode sponge structure
Variables
-
character(len=40) isomip_initialization::mdl = "ISOMIP_initialization" This module’s name.
-
-
namespace
isomip_tracer¶ Routines used to set up and use a set of (one for now) dynamically passive tracers in the ISOMIP configuration.
For now, just one passive tracer is injected in the sponge layer.
Functions
-
logical function, public isomip_tracer::register_isomip_tracer(HI HI, GV GV, param_file param_file, CS CS, tr_Reg tr_Reg, restart_CS restart_CS) This subroutine is used to register tracer fields.
- Parameters
[in] hi: A horizontal index type structure.[in] gv: The ocean’s vertical grid structure.[in] param_file: A structure indicating the open file to parse for model parameter values.cs: A pointer that is set to point to the control structure for this module (in/out).tr_reg: A pointer to the tracer registry.restart_cs: A pointer to the restart control structure.
-
subroutine, public isomip_tracer::initialize_isomip_tracer(restart restart, day day, G G, GV GV, h h, diag diag, OBC OBC, CS CS, ALE_sponge_CSp ALE_sponge_CSp) Initializes the NTR tracer fields in tr(:,:,:,:)
- Parameters
[in] g: Grid structure.[in] gv: The ocean’s vertical grid structure.[in] restart: .true. if the fields have already been read from a restart file.[in] day: Time of the start of the run.[in] h: Layer thickness [H ~> m or kg m-2].[in] diag: A structure that is used to regulate diagnostic output.obc: This open boundary condition type specifies whether, where, and what open boundary conditions are used. This is not being used for now.cs: The control structure returned by a previous call to ISOMIP_register_tracer.ale_sponge_csp: A pointer to the control structure for the sponges, if they are in use. Otherwise this may be unassociated.
-
subroutine, public isomip_tracer::isomip_tracer_column_physics(h_old h_old, h_new h_new, ea ea, eb eb, fluxes fluxes, dt dt, G G, GV GV, CS CS, evap_CFL_limit evap_CFL_limit, minimum_forcing_depth minimum_forcing_depth) This subroutine applies diapycnal diffusion, including the surface boundary conditions and any other column tracer physics or chemistry to the tracers from this file.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h_old: Layer thickness before entrainment [H ~> m or kg m-2].[in] h_new: Layer thickness after entrainment [H ~> m or kg m-2].[in] ea: an array to which the amount of fluid entrained[in] eb: an array to which the amount of fluid entrained[in] fluxes: A structure containing pointers to thermodynamic and tracer forcing fields. Unused fields have NULL ptrs.[in] dt: The amount of time covered by this call [s]cs: The control structure returned by a previous call to ISOMIP_register_tracer.[in] evap_cfl_limit: Limit on the fraction of the water that can be fluxed out of the top layer in a timestep [nondim][in] minimum_forcing_depth: The smallest depth over which fluxes can be applied [m]
-
subroutine, public isomip_tracer::isomip_tracer_surface_state(state state, h h, G G, CS CS) This subroutine extracts the surface fields from this tracer package that are to be shared with the atmosphere in coupled configurations. This particular tracer package does not report anything back to the coupler.
- Parameters
[in] g: The ocean’s grid structure.[inout] state: A structure containing fields that describe the surface state of the ocean.[in] h: Layer thickness [H ~> m or kg m-2].cs: The control structure returned by a previous call to ISOMIP_register_tracer.
-
subroutine, public isomip_tracer::isomip_tracer_end(CS CS) Deallocate any memory used by the ISOMIP tracer package.
- Parameters
cs: The control structure returned by a previous call to ISOMIP_register_tracer.
Variables
-
integer, parameter isomip_tracer::ntr = 1 ntr is the number of tracers in this module.
-
-
namespace
kelvin_initialization¶ Configures the model for the Kelvin wave experiment.
Kelvin = coastally-trapped Kelvin waves from the ROMS examples. Initialize with level surfaces and drive the wave in at the west, radiate out at the east.
Functions
-
logical function, public kelvin_initialization::register_kelvin_obc(param_file param_file, CS CS, OBC_Reg OBC_Reg) Add Kelvin wave to OBC registry.
- Parameters
[in] param_file: parameter file.cs: Kelvin wave control structure.obc_reg: OBC registry.
-
subroutine, public kelvin_initialization::kelvin_obc_end(CS CS) Clean up the Kelvin wave OBC from registry.
- Parameters
cs: Kelvin wave control structure.
-
subroutine, public kelvin_initialization::kelvin_initialize_topography(D D, G G, param_file param_file, max_depth max_depth, US US) This subroutine sets up the Kelvin topography and land mask.
- Parameters
[in] g: The dynamic horizontal grid type[out] d: Ocean bottom depth in m or Z if US is present[in] param_file: Parameter file structure[in] max_depth: Maximum model depth in the units of D[in] us: A dimensional unit scaling type
-
subroutine, public kelvin_initialization::kelvin_set_obc_data(OBC OBC, CS CS, G G, GV GV, US US, h h, Time Time) This subroutine sets the properties of flow at open boundary conditions.
- Parameters
obc: This open boundary condition type specifies whether, where, and what open boundary conditions are used.cs: Kelvin wave control structure.[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] h: layer thickness [H ~> m or kg m-2].[in] time: model time.
-
-
namespace
lock_exchange_initialization¶ Initialization of the “lock exchange” experiment. lock_exchange = A 2-d density driven hydraulic exchange flow.
Functions
-
subroutine, public lock_exchange_initialization::lock_exchange_initialize_thickness(h h, G G, GV GV, US US, param_file param_file, just_read_params just_read_params) This subroutine initializes layer thicknesses for the lock_exchange experiment.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[out] h: The thickness that is being initialized [H ~> m or kg m-2].[in] param_file: A structure indicating the open file to parse for model parameter values.[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
-
namespace
meso_surface_forcing¶ Sets forcing for the MESO configuration.
Rewritten by Robert Hallberg, June 2009
This file contains the subroutines that a user should modify to to set the surface wind stresses and fluxes of buoyancy or temperature and fresh water. They are called when the run-time parameters WIND_CONFIG or BUOY_CONFIG are set to “USER”. The standard version has simple examples, along with run-time error messages that will cause the model to abort if this code has not been modified. This code is intended for use with relatively simple specifications of the forcing. For more complicated forms, it is probably a good idea to read the forcing from input files using “file” for WIND_CONFIG and BUOY_CONFIG.
MESO_buoyancy forcing is used to set the surface buoyancy forcing, which may include a number of fresh water flux fields (evap, liq_precip, froz_precip, liq_runoff, froz_runoff, and vprec) and the surface heat fluxes (sw, lw, latent and sens) if temperature and salinity are state variables, or it may simply be the buoyancy flux if it is not. This routine also has coded a restoring to surface values of temperature and salinity.
Functions
-
subroutine, public meso_surface_forcing::meso_buoyancy_forcing(sfc_state sfc_state, fluxes fluxes, day day, dt dt, G G, US US, CS CS) This subroutine sets up the MESO buoyancy forcing, which uses control-theory style specification restorative buoyancy fluxes at large scales.
- Parameters
[inout] sfc_state: A structure containing fields that describe the surface state of the ocean.[inout] fluxes: A structure containing thermodynamic forcing fields[in] day: The time of the fluxes[in] dt: The amount of time over which the fluxes apply [s][in] g: The ocean’s grid structure[in] us: A dimensional unit scaling typecs: A pointer to the control structure returned by a previous call to MESO_surface_forcing_init
-
subroutine, public meso_surface_forcing::meso_surface_forcing_init(Time Time, G G, US US, param_file param_file, diag diag, CS CS) Initialize the MESO surface forcing module.
- Parameters
[in] time: The current model time[in] g: The ocean’s grid structure[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters[inout] diag: structure used to regulate diagnostic outputcs: A pointer that is set to point to the control structure for this module
Variables
-
logical
first_call= .true.¶ True until after the first call to the MESO forcing routines.
-
-
namespace
midas_vertmap¶ Routines for initialization callable from MOM6 or Python (MIDAS)
Functions
-
real function, dimension(size(tr_in,1),size(tr_in,2),nlay), public midas_vertmap::tracer_z_init(tr_in tr_in, z_edges z_edges, e e, nkml nkml, nkbl nkbl, land_fill land_fill, wet wet, nlay nlay, nlevs nlevs, debug debug, i_debug i_debug, j_debug j_debug, eps_z eps_z) Layer model routine for remapping tracers.
- Return
tracers in layer space
- Parameters
[in] tr_in: The z-space array of tracer concentrations that is read in.[in] z_edges: The depths of the cell edges in the input z* data [Z ~> m or m][in] nlay: The number of vertical layers in the target grid[in] e: The depths of the target layer interfaces [Z ~> m or m][in] nkml: The number of mixed layers[in] nkbl: The number of buffer layers[in] land_fill: fill in data over land (1)[in] wet: The wet mask for the source data (valid points)[in] nlevs: The number of input levels with valid data[in] debug: optional debug flag[in] i_debug: i-index of point for debugging[in] j_debug: j-index of point for debugging[in] eps_z: A negligibly small layer thickness [Z ~> m or m].
-
integer function, dimension(size(a,1),size(x,1)) midas_vertmap::bisect_fast(a a, x x, lo lo, hi hi) Return the index where to insert item x in list a, assuming a is sorted. The return values [i] is such that all e in a[:i-1] have e <= x, and all e in a[i:] have e > x. So if x already appears in the list, will insert just after the rightmost x already there. Optional args lo (default 1) and hi (default len(a)) bound the slice of a to be searched.
- Parameters
[in] a: Sorted list[in] x: Item to be inserted[in] lo: Lower bracket of optional range to search[in] hi: Upper bracket of optional range to search
-
subroutine, public midas_vertmap::determine_temperature(temp temp, salt salt, R R, p_ref p_ref, niter niter, land_fill land_fill, h h, k_start k_start, eos eos) This subroutine determines the potential temperature and salinity that is consistent with the target density using provided initial guess.
- Parameters
[inout] temp: potential temperature [degC][inout] salt: salinity [PSU][in] r: desired potential density [kg m-3].[in] p_ref: reference pressure [Pa].[in] niter: maximum number of iterations[in] k_start: starting index (i.e. below the buffer layer)[in] land_fill: land fill value[in] h: layer thickness, used only to avoid working on massless layerseos: seawater equation of state control structure
-
subroutine
find_overlap(e e, Z_top Z_top, Z_bot Z_bot, k_max k_max, k_start k_start, k_top k_top, k_bot k_bot, wt wt, z1 z1, z2 z2)¶ This subroutine determines the layers bounded by interfaces e that overlap with the depth range between Z_top and Z_bot, and also the fractional weights of each layer. It also calculates the normalized relative depths of the range of each layer that overlaps that depth range. Note that by convention, e decreases with increasing k and Z_top > Z_bot.
- Parameters
[in] e: The interface positions, [Z ~> m] or other units.[in] z_top: The top of the range being mapped to, [Z ~> m] or other units.[in] z_bot: The bottom of the range being mapped to, [Z ~> m] or other units.[in] k_max: The number of valid layers.[in] k_start: The layer at which to start searching.[out] k_top: The index of the top layer that overlap with the depth range.[out] k_bot: The index of the bottom layer that overlap with the depth range.[out] wt: The relative weights of each layer from k_top to k_bot [nondim].[out] z1: Depth of the top limit of layer that contributes to a level [nondim].[out] z2: Depth of the bottom limit of layer that contributes to a level [nondim].
-
real function midas_vertmap::find_limited_slope(val val, e e, k k) This subroutine determines a limited slope for val to be advected with a piecewise limited scheme.
- Return
The normalized slope in the intracell distribution of val.
- Parameters
[in] val: An column the values that are being interpolated.[in] e: A column’s interface heights [Z ~> m] or other units.[in] k: The layer whose slope is being determined.
-
real function, dimension(size(rho,1),size(rho,2),size(rb,1)), public midas_vertmap::find_interfaces(rho rho, zin zin, Rb Rb, depth depth, nlevs nlevs, nkml nkml, nkbl nkbl, hml hml, debug debug, eps_z eps_z) Find interface positions corresponding to density profile.
- Return
The returned interface, in the same units az zin.
- Parameters
[in] rho: potential density in z-space [kg m-3][in] zin: Input data levels [Z ~> m or m].[in] rb: target interface densities [kg m-3][in] depth: ocean depth [Z ~> m].[in] nlevs: number of valid points in each column[in] debug: optional debug flag[in] nkml: number of mixed layer pieces[in] nkbl: number of buffer layer pieces[in] hml: mixed layer depth [Z ~> m].[in] eps_z: A negligibly small layer thickness [Z ~> m or m].
-
subroutine, public midas_vertmap::meshgrid(x x, y y, x_T x_T, y_T y_T) Create a 2d-mesh of grid coordinates from 1-d arrays.
- Parameters
[in] x: input x coordinates[in] y: input y coordinates[inout] x_t: output 2-d version[inout] y_t: output 2-d version
-
subroutine
smooth_heights(zi zi, fill fill, bad bad, sor sor, niter niter, cyclic_x cyclic_x, tripolar_n tripolar_n)¶ Solve del2 (zi) = 0 using successive iterations with a 5 point stencil. Only points fill==1 are modified. Except where bad==1, information propagates isotropically in index space. The resulting solution in each region is an approximation to del2(zi)=0 subject to boundary conditions along the valid points curve bounding this region.
- Parameters
[inout] zi: interface positions [m] or arbitrary[in] fill: points to be smoothed[in] bad: ignore these points[in] sor: successive over-relaxation coefficient (typically 0.6)[in] niter: maximum number of iterations[in] cyclic_x: input grid cyclic condition in the zonal direction[in] tripolar_n: tripolar Arctic fold flag
-
integer function, dimension(0:size(m,1)+1,0:size(m,2)+1) midas_vertmap::fill_boundaries_int(m m, cyclic_x cyclic_x, tripolar_n tripolar_n) Fill grid edges.
- Return
output filled array
- Parameters
[in] m: input array[in] cyclic_x: zonal cyclic condition[in] tripolar_n: northern fold condition
-
real function, dimension(0:size(m,1)+1,0:size(m,2)+1) midas_vertmap::fill_boundaries_real(m m, cyclic_x cyclic_x, tripolar_n tripolar_n) fill grid edges
- Return
output filled array
- Parameters
[in] m: input array[in] cyclic_x: zonal cyclic condition[in] tripolar_n: northern fold condition
-
-
namespace
mom¶ The central module of the MOM6 ocean model.
Modular Ocean Model (MOM) Version 6.0 (MOM6)
Additional contributions from:
Whit Anderson
Brian Arbic
Will Cooke
Anand Gnanadesikan
Matthew Harrison
Mehmet Ilicak
Laura Jackson
Jasmine John
John Krasting
Zhi Liang
Bonnie Samuels
Harper Simmons
Laurent White
Niki Zadeh
- Authors
Alistair Adcroft, Robert Hallberg, and Stephen Griffies
MOM ice-shelf code was developed by
Daniel Goldberg
Robert Hallberg
Chris Little
Olga Sergienko
Unnamed Group
-
integer
id_clock_ocean¶ CPU time clock IDs.
-
integer
id_clock_dynamics¶ CPU time clock IDs.
-
integer
id_clock_thermo¶ CPU time clock IDs.
-
integer
id_clock_tracer¶ CPU time clock IDs.
-
integer
id_clock_diabatic¶ CPU time clock IDs.
-
integer
id_clock_continuity¶ CPU time clock IDs.
-
integer
id_clock_thick_diff¶ CPU time clock IDs.
-
integer
id_clock_bbl_visc¶ CPU time clock IDs.
-
integer
id_clock_ml_restrat¶ CPU time clock IDs.
-
integer
id_clock_diagnostics¶ CPU time clock IDs.
-
integer
id_clock_z_diag¶ CPU time clock IDs.
-
integer
id_clock_init¶ CPU time clock IDs.
-
integer
id_clock_mom_init¶ CPU time clock IDs.
-
integer
id_clock_pass¶ CPU time clock IDs.
-
integer
id_clock_pass_init¶ CPU time clock IDs.
-
integer
id_clock_ale¶ CPU time clock IDs.
-
integer
id_clock_other¶ CPU time clock IDs.
-
integer
id_clock_offline_tracer¶ CPU time clock IDs.
-
subroutine, public mom::step_mom(forces forces, fluxes fluxes, sfc_state sfc_state, Time_start Time_start, time_interval time_interval, CS CS, Waves Waves, do_dynamics do_dynamics, do_thermodynamics do_thermodynamics, start_cycle start_cycle, end_cycle end_cycle, cycle_length cycle_length, reset_therm reset_therm) This subroutine orchestrates the time stepping of MOM. The adiabatic dynamics are stepped by calls to one of the step_MOM_dyn_…routines. The action of lateral processes on tracers occur in calls to advect_tracer and tracer_hordiff. Vertical mixing and possibly remapping occur inside of diabatic.
- Parameters
[inout] forces: A structure with the driving mechanical forces[inout] fluxes: A structure with pointers to themodynamic, tracer and mass exchange forcing fields[inout] sfc_state: surface ocean state[in] time_start: starting time of a segment, as a time type[in] time_interval: time interval covered by this run segment [s].cs: control structure from initialize_MOMwaves: An optional pointer to a wave property CS[in] do_dynamics: Present and false, do not do updates due to the dynamics.[in] do_thermodynamics: Present and false, do not do updates due to the thermodynamics or remapping.[in] start_cycle: This indicates whether this call is to be treated as the first call to step_MOM in a time-stepping cycle; missing is like true.[in] end_cycle: This indicates whether this call is to be treated as the last call to step_MOM in a time-stepping cycle; missing is like true.[in] cycle_length: The amount of time in a coupled time stepping cycle [s].[in] reset_therm: This indicates whether the running sums of thermodynamic quantities should be reset. If missing, this is like start_cycle.
-
subroutine
step_mom_dynamics(forces forces, p_surf_begin p_surf_begin, p_surf_end p_surf_end, dt dt, dt_thermo dt_thermo, bbl_time_int bbl_time_int, CS CS, Time_local Time_local, Waves Waves)¶ Time step the ocean dynamics, including the momentum and continuity equations.
- Parameters
[in] forces: A structure with the driving mechanical forcesp_surf_begin: A pointer (perhaps NULL) to the surface pressure at the beginning of this dynamic step, intent in [Pa].p_surf_end: A pointer (perhaps NULL) to the surface pressure at the end of this dynamic step, intent in [Pa].[in] dt: time interval covered by this call [s].[in] dt_thermo: time interval covered by any updates that may span multiple dynamics steps [s].[in] bbl_time_int: time interval over which updates to the bottom boundary layer properties will apply [s], or zero not to update the properties.cs: control structure from initialize_MOM[in] time_local: End time of a segment, as a time typewaves: Container for wave related parameters; the
-
subroutine
step_mom_tracer_dyn(CS CS, G G, GV GV, h h, Time_local Time_local)¶ step_MOM_tracer_dyn does tracer advection and lateral diffusion, bringing the tracers up to date with the changes in state due to the dynamics. Surface sources and sinks and remapping are handled via step_MOM_thermo.
- Parameters
[inout] cs: control structure[inout] g: ocean grid structure[in] gv: ocean vertical grid structure[in] h: layer thicknesses after the transports [H ~> m or kg m-2][in] time_local: The model time at the end of the time step.
-
subroutine
step_mom_thermo(CS CS, G G, GV GV, US US, u u, v v, h h, tv tv, fluxes fluxes, dtdia dtdia, Time_end_thermo Time_end_thermo, update_BBL update_BBL, Waves Waves)¶ MOM_step_thermo orchestrates the thermodynamic time stepping and vertical remapping, via calls to diabatic (or adiabatic) and ALE_main.
- Parameters
[inout] cs: Master MOM control structure[inout] g: ocean grid structure[inout] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] u: zonal velocity [m s-1][inout] v: meridional velocity [m s-1][inout] h: layer thickness [H ~> m or kg m-2][inout] tv: A structure pointing to various thermodynamic variables[inout] fluxes: pointers to forcing fields[in] dtdia: The time interval over which to advance [s][in] time_end_thermo: End of averaging interval for thermo diags[in] update_bbl: If true, calculate the bottom boundary layer properties.waves: Container for wave related parameters
-
subroutine, public mom::step_offline(forces forces, fluxes fluxes, sfc_state sfc_state, Time_start Time_start, time_interval time_interval, CS CS) step_offline is the main driver for running tracers offline in MOM6. This has been primarily developed with ALE configurations in mind. Some work has been done in isopycnal configuration, but the work is very preliminary. Some more detail about this capability along with some of the subroutines called here can be found in tracers/MOM_offline_control.F90
- Parameters
[in] forces: A structure with the driving mechanical forces[inout] fluxes: pointers to forcing fields[inout] sfc_state: surface ocean state[in] time_start: starting time of a segment, as a time type[in] time_interval: time intervalcs: control structure from initialize_MOM
-
subroutine, public mom::initialize_mom(Time Time, Time_init Time_init, param_file param_file, dirs dirs, CS CS, restart_CSp restart_CSp, Time_in Time_in, offline_tracer_mode offline_tracer_mode, input_restart_file input_restart_file, diag_ptr diag_ptr, count_calls count_calls, tracer_flow_CSp tracer_flow_CSp) Initialize MOM, including memory allocation, setting up parameters and diagnostics, initializing the ocean state variables, and initializing subsidiary modules.
- Parameters
[inout] time: model time, set in this routine[in] time_init: The start time for the coupled model’s calendar[out] param_file: structure indicating parameter file to parse[out] dirs: structure with directory pathscs: pointer set in this routine to MOM control structurerestart_csp: pointer set in this routine to the restart control structure that will be used for MOM.[in] time_in: time passed to MOM_initialize_state when model is not being started from a restart file[out] offline_tracer_mode: True is returned if tracers are being run offline[in] input_restart_file: If present, name of restart file to readdiag_ptr: A pointer set in this routine to the diagnostic regulatory structuretracer_flow_csp: A pointer set in this routine to[in] count_calls: If true, nstep_tot counts the number of calls to step_MOM instead of the number of dynamics timesteps.
-
subroutine, public mom::finish_mom_initialization(Time Time, dirs dirs, CS CS, restart_CSp restart_CSp) Finishes initializing MOM and writes out the initial conditions.
- Parameters
[in] time: model time, used in this routine[in] dirs: structure with directory pathscs: pointer to MOM control structurerestart_csp: pointer to the restart control structure that will be used for MOM.
-
subroutine
register_diags(Time Time, G G, GV GV, IDs IDs, diag diag)¶ Register certain diagnostics.
- Parameters
[in] time: current model time[in] g: ocean grid structure[in] gv: ocean vertical grid structure[inout] ids: A structure with the diagnostic IDs.[inout] diag: regulates diagnostic output
-
subroutine
mom_timing_init(CS CS)¶ Set up CPU clock IDs for timing various subroutines.
- Parameters
[in] cs: control structure set up by initialize_MOM.
-
subroutine
set_restart_fields(GV GV, US US, param_file param_file, CS CS, restart_CSp restart_CSp)¶ Set the fields that are needed for bitwise identical restarting the time stepping scheme. In addition to those specified here directly, there may be fields related to the forcing or to the barotropic solver that are needed; these are specified in sub- routines that are called from this one.
This routine should be altered if there are any changes to the time stepping scheme. The CHECK_RESTART facility may be used to confirm that all needed restart fields have been included.
- Parameters
[inout] gv: ocean vertical grid structure[inout] us: A dimensional unit scaling type[in] param_file: opened file for parsing to get parameters[in] cs: control structure set up by inialize_MOMrestart_csp: pointer to the restart control structure that will be used for MOM.
-
subroutine
adjust_ssh_for_p_atm(tv tv, G G, GV GV, US US, ssh ssh, p_atm p_atm, use_EOS use_EOS)¶ Apply a correction to the sea surface height to compensate for the atmospheric pressure (the inverse barometer).
- Parameters
[in] tv: A structure pointing to various thermodynamic variables[in] g: ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] ssh: time mean surface height [m]p_atm: atmospheric pressure [Pa][in] use_eos: If true, calculate the density for the SSH correction using the equation of state.
-
subroutine, public mom::extract_surface_state(CS CS, sfc_state sfc_state) Set the surface (return) properties of the ocean model by setting the appropriate fields in sfc_state. Unused fields are set to NULL or are unallocated.
- Parameters
cs: Master MOM control structure[inout] sfc_state: transparent ocean surface state structure shared with the calling routine data in this structure is intent out.
-
logical function, public mom::mom_state_is_synchronized(CS CS, adv_dyn adv_dyn) Return true if all phases of step_MOM are at the same point in time.
- Return
True if all phases of the update are synchronized.
- Parameters
cs: MOM control structure[in] adv_dyn: If present and true, only check whether the advection is up-to-date with the dynamics.
-
subroutine, public mom::get_mom_state_elements(CS CS, G G, GV GV, US US, C_p C_p, use_temp use_temp) This subroutine offers access to values or pointers to other types from within the MOM_control_struct, allowing the MOM_control_struct to be opaque.
- Parameters
cs: MOM control structureg: structure containing metrics and grid infogv: structure containing vertical grid infous: A dimensional unit scaling type[out] c_p: The heat capacity[out] use_temp: Indicates whether temperature is a state variable
-
subroutine, public mom::get_ocean_stocks(CS CS, mass mass, heat heat, salt salt, on_PE_only on_PE_only) Find the global integrals of various quantities.
- Parameters
cs: MOM control structure[out] heat: The globally integrated integrated ocean heat [J].[out] salt: The globally integrated integrated ocean salt [kg].[out] mass: The globally integrated integrated ocean mass [kg].[in] on_pe_only: If present and true, only sum on the local PE.
-
subroutine, public mom::mom_end(CS CS) End of ocean model, including memory deallocation.
- Parameters
cs: MOM control structure
-
namespace
mom_ale¶ This module contains the main regridding routines.
Regridding comprises two steps:
Interpolation and creation of a new grid based on target interface densities (or any other criterion).
Remapping of quantities between old grid and new grid.
Original module written by Laurent White, 2008.06.09
Functions
-
subroutine, public mom_ale::ale_init(param_file param_file, GV GV, US US, max_depth max_depth, CS CS) This routine is typically called (from initialize_MOM in file MOM.F90) before the main time integration loop to initialize the regridding stuff. We read the MOM_input file to register the values of different regridding/remapping parameters.
- Parameters
[in] param_file: Parameter file[in] gv: Ocean vertical grid structure[in] us: A dimensional unit scaling type[in] max_depth: The maximum depth of the ocean [Z ~> m].cs: Module control structure
-
subroutine, public mom_ale::ale_register_diags(Time Time, G G, GV GV, US US, diag diag, CS CS) Initialize diagnostics for the ALE module.
- Parameters
[in] time: Time structure[in] g: Grid structure[in] us: A dimensional unit scaling type[in] gv: Ocean vertical grid structure[in] diag: Diagnostics control structurecs: Module control structure
-
subroutine, public mom_ale::adjustgridforintegrity(CS CS, G G, GV GV, h h) Crudely adjust (initial) grid for integrity. This routine is typically called (from initialize_MOM in file MOM.F90) before the main time integration loop to initialize the regridding stuff. We read the MOM_input file to register the values of different regridding/remapping parameters.
- Parameters
cs: Regridding parameters and options[in] g: Ocean grid informations[in] gv: Ocean vertical grid structure[inout] h: Current 3D grid thickness that are to be adjusted [H ~> m or kg-2]
-
subroutine, public mom_ale::ale_end(CS CS) End of regridding (memory deallocation). This routine is typically called (from MOM_end in file MOM.F90) after the main time integration loop to deallocate the regridding stuff.
- Parameters
cs: module control structure
-
subroutine, public mom_ale::ale_main(G G, GV GV, US US, h h, u u, v v, tv tv, Reg Reg, CS CS, dt dt, frac_shelf_h frac_shelf_h) Takes care of (1) building a new grid and (2) remapping all variables between the old grid and the new grid. The creation of the new grid can be based on z coordinates, target interface densities, sigma coordinates or any arbitrary coordinate system.
- Parameters
[in] g: Ocean grid informations[in] gv: Ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] h: Current 3D grid obtained after the last time step [H ~> m or kg m-2][inout] u: Zonal velocity field [m s-1][inout] v: Meridional velocity field [m s-1][inout] tv: Thermodynamic variable structurereg: Tracer registry structurecs: Regridding parameters and options[in] dt: Time step between calls to ALE_main()frac_shelf_h: Fractional ice shelf coverage
-
subroutine, public mom_ale::ale_main_offline(G G, GV GV, h h, tv tv, Reg Reg, CS CS, dt dt) Takes care of (1) building a new grid and (2) remapping all variables between the old grid and the new grid. The creation of the new grid can be based on z coordinates, target interface densities, sigma coordinates or any arbitrary coordinate system.
- Parameters
[in] g: Ocean grid informations[in] gv: Ocean vertical grid structure[inout] h: Current 3D grid obtained after the last time step [H ~> m or kg-2][inout] tv: Thermodynamic variable structurereg: Tracer registry structurecs: Regridding parameters and options[in] dt: Time step between calls to ALE_main()
-
subroutine, public mom_ale::ale_offline_inputs(CS CS, G G, GV GV, h h, tv tv, Reg Reg, uhtr uhtr, vhtr vhtr, Kd Kd, debug debug) Regrid/remap stored fields used for offline tracer integrations. These input fields are assumed to have the same layer thicknesses at the end of the last offline interval (which should be a Zstar grid). This routine builds a grid on the runtime specified vertical coordinate.
- Parameters
cs: Regridding parameters and options[in] g: Ocean grid informations[in] gv: Ocean vertical grid structure[inout] h: Layer thicknesses[inout] tv: Thermodynamic variable structurereg: Tracer registry structure[inout] uhtr: Zonal mass fluxes[inout] vhtr: Meridional mass fluxes[inout] kd: Input diffusivites[in] debug: If true, then turn checksums
-
subroutine, public mom_ale::ale_offline_tracer_final(G G, GV GV, h h, tv tv, h_target h_target, Reg Reg, CS CS) Remaps all tracers from h onto h_target. This is intended to be called when tracers are done offline. In the case where transports don’t quite conserve, we still want to make sure that layer thicknesses offline do not drift too far away from the online model.
- Parameters
[in] g: Ocean grid informations[in] gv: Ocean vertical grid structure[inout] h: Current 3D grid obtained after the last time step [H ~> m or kg-2][inout] tv: Thermodynamic variable structure[inout] h_target: Current 3D grid obtained after last time step [H ~> m or kg-2]reg: Tracer registry structurecs: Regridding parameters and options
-
subroutine
check_grid(G G, GV GV, h h, threshold threshold)¶ Check grid for negative thicknesses.
- Parameters
[in] g: Ocean grid structure[in] gv: Ocean vertical grid structure[in] h: Current 3D grid obtained after the last time step [H ~> m or kg m-2][in] threshold: Value below which to flag issues, [H ~> m or kg m-2]
-
subroutine, public mom_ale::ale_build_grid(G G, GV GV, regridCS regridCS, remapCS remapCS, h h, tv tv, debug debug, frac_shelf_h frac_shelf_h) Generates new grid.
- Parameters
[in] g: Ocean grid structure[in] gv: Ocean vertical grid structure[in] regridcs: Regridding parameters and options[in] remapcs: Remapping parameters and options[inout] tv: Thermodynamical variable structure[inout] h: Current 3D grid obtained after the last time step [H ~> m or kg-2][in] debug: If true, show the call treefrac_shelf_h: Fractional ice shelf coverage
-
subroutine, public mom_ale::ale_regrid_accelerated(CS CS, G G, GV GV, h h, tv tv, n n, u u, v v, Reg Reg, dt dt, dzRegrid dzRegrid, initial initial) For a state-based coordinate, accelerate the process of regridding by repeatedly applying the grid calculation algorithm.
- Parameters
cs: ALE control structure[inout] g: Ocean grid[in] gv: Vertical grid[inout] h: Original thicknesses[inout] tv: Thermo vars (T/S/EOS)[in] n: Number of times to regrid[inout] u: Zonal velocity[inout] v: Meridional velocityreg: Tracer registry to remap onto new grid[in] dt: Model timestep to provide a timescale for regridding[inout] dzregrid: Final change in interface positions[in] initial: Whether we’re being called from an initialization routine (and expect diagnostics to work)
-
subroutine
remap_all_state_vars(CS_remapping CS_remapping, CS_ALE CS_ALE, G G, GV GV, h_old h_old, h_new h_new, Reg Reg, dxInterface dxInterface, u u, v v, debug debug, dt dt)¶ This routine takes care of remapping all variable between the old and the new grids. When velocity components need to be remapped, thicknesses at velocity points are taken to be arithmetic averages of tracer thicknesses. This routine is called during initialization of the model at time=0, to remap initiali conditions to the model grid. It is also called during a time step to update the state.
- Parameters
[in] cs_remapping: Remapping control structure[in] cs_ale: ALE control structure[in] g: Ocean grid structure[in] gv: Ocean vertical grid structure[in] h_old: Thickness of source grid [H ~> m or kg-2][in] h_new: Thickness of destination grid [H ~> m or kg-2]reg: Tracer registry structure[in] dxinterface: Change in interface position[inout] u: Zonal velocity component [m s-1][inout] v: Meridional velocity component [m s-1][in] debug: If true, show the call tree[in] dt: time step for diagnostics
-
subroutine, public mom_ale::ale_remap_scalar(CS CS, G G, GV GV, nk_src nk_src, h_src h_src, s_src s_src, h_dst h_dst, s_dst s_dst, all_cells all_cells, old_remap old_remap) Remaps a single scalar between grids described by thicknesses h_src and h_dst. h_dst must be dimensioned as a model array with GVke layers while h_src can have an arbitrary number of layers specified by nk_src.
- Parameters
[in] cs: Remapping control structure[in] g: Ocean grid structure[in] gv: Ocean vertical grid structure[in] nk_src: Number of levels on source grid[in] h_src: Level thickness of source grid [H ~> m or kg-2][in] s_src: Scalar on source grid[in] h_dst: Level thickness of destination grid [H ~> m or kg-2][inout] s_dst: Scalar on destination grid[in] all_cells: If false, only reconstruct for non-vanished cells. Use all vanished layers otherwise (default).[in] old_remap: If true, use the old “remapping_core_w” method, otherwise use “remapping_core_h”.
-
subroutine, public mom_ale::pressure_gradient_plm(CS CS, S_t S_t, S_b S_b, T_t T_t, T_b T_b, G G, GV GV, tv tv, h h, bdry_extrap bdry_extrap) Use plm reconstruction for pressure gradient (determine edge values) By using a PLM (limited piecewise linear method) reconstruction, this routine determines the edge values for the salinity and temperature within each layer. These edge values are returned and are used to compute the pressure gradient (by computing the densities).
- Parameters
[in] g: ocean grid structure[in] gv: Ocean vertical grid structure[inout] cs: module control structure[inout] s_t: Salinity at the top edge of each layer[inout] s_b: Salinity at the bottom edge of each layer[inout] t_t: Temperature at the top edge of each layer[inout] t_b: Temperature at the bottom edge of each layer[in] tv: thermodynamics structure[in] h: layer thickness [H ~> m or kg m-2][in] bdry_extrap: If true, use high-order boundary extrapolation within boundary cells
-
subroutine, public mom_ale::pressure_gradient_ppm(CS CS, S_t S_t, S_b S_b, T_t T_t, T_b T_b, G G, GV GV, tv tv, h h, bdry_extrap bdry_extrap) Use ppm reconstruction for pressure gradient (determine edge values) By using a PPM (limited piecewise linear method) reconstruction, this routine determines the edge values for the salinity and temperature within each layer. These edge values are returned and are used to compute the pressure gradient (by computing the densities).
- Parameters
[in] g: ocean grid structure[in] gv: Ocean vertical grid structure[inout] cs: module control structure[inout] s_t: Salinity at the top edge of each layer[inout] s_b: Salinity at the bottom edge of each layer[inout] t_t: Temperature at the top edge of each layer[inout] t_b: Temperature at the bottom edge of each layer[in] tv: thermodynamics structure[in] h: layer thicknesses [H ~> m or kg m-2][in] bdry_extrap: If true, use high-order boundary extrapolation within boundary cells
-
subroutine, public mom_ale::ale_initregridding(GV GV, US US, max_depth max_depth, param_file param_file, mdl mdl, regridCS regridCS) Initializes regridding for the main ALE algorithm.
- Parameters
[in] gv: Ocean vertical grid structure[in] us: A dimensional unit scaling type[in] max_depth: The maximum depth of the ocean [Z ~> m].[in] param_file: parameter file[in] mdl: Name of calling module[out] regridcs: Regridding parameters and work arrays
-
real function, dimension(cs%nk+1), public mom_ale::ale_getcoordinate(CS CS) Query the target coordinate interfaces positions.
- Parameters
cs: module control structure
-
character(len=20) function, public mom_ale::ale_getcoordinateunits(CS CS) Query the target coordinate units.
- Parameters
cs: module control structure
-
logical function, public mom_ale::ale_remap_init_conds(CS CS) Returns true if initial conditions should be regridded and remapped.
- Parameters
cs: module control structure
-
subroutine, public mom_ale::ale_update_regrid_weights(dt dt, CS CS) Updates the weights for time filtering the new grid generated in regridding.
- Parameters
[in] dt: Time-step used between ALE callscs: ALE control structure
-
subroutine, public mom_ale::ale_updateverticalgridtype(CS CS, GV GV) Update the vertical grid type with ALE information. This subroutine sets information in the verticalGrid_type to be consistent with the use of ALE mode.
- Parameters
cs: ALE control structuregv: vertical grid information
-
subroutine, public mom_ale::ale_writecoordinatefile(CS CS, GV GV, directory directory) Write the vertical coordinate information into a file. This subroutine writes out a file containing any available data related to the vertical grid used by the MOM ocean model when in ALE mode.
- Parameters
cs: module control structure[in] gv: ocean vertical grid structure[in] directory: directory for writing grid info
-
subroutine, public mom_ale::ale_initthicknesstocoord(CS CS, G G, GV GV, h h) Set h to coordinate values for fixed coordinate systems.
- Parameters
[inout] cs: module control structure[in] g: module grid structure[in] gv: Ocean vertical grid structure[out] h: layer thickness [H ~> m or kg m-2]
-
namespace
mom_ale_sponge¶ This module contains the routines used to apply sponge layers when using the ALE mode.
Applying sponges requires the following:
initialize_ALE_sponge
set_up_ALE_sponge_field (tracers) and set_up_ALE_sponge_vel_field (vel)
apply_ALE_sponge
init_ALE_sponge_diags (not being used for now)
ALE_sponge_end (not being used for now)
Functions
-
subroutine
initialize_ale_sponge_fixed(Iresttime Iresttime, G G, param_file param_file, CS CS, data_h data_h, nz_data nz_data)¶ This subroutine determines the number of points which are within sponges in this computational domain. Only points that have positive values of Iresttime and which mask2dT indicates are ocean points are included in the sponges. It also stores the target interface heights.
- Parameters
[in] g: The ocean’s grid structure.[in] nz_data: The total number of sponge input layers.[in] iresttime: The inverse of the restoring time [s-1].[in] param_file: A structure indicating the open file to parse for model parameter values.cs: A pointer that is set to point to the control structure for this module (in/out).[in] data_h: The thicknesses of the sponge input layers [H ~> m or kg m-2].
-
integer function, public mom_ale_sponge::get_ale_sponge_nz_data(CS CS) Return the number of layers in the data with a fixed ALE sponge, or 0 if there are no sponge columns on this PE.
- Return
The number of layers in the fixed sponge data.
- Parameters
cs: A pointer that is set to point to the control structure for the ALE_sponge module.
-
subroutine, public mom_ale_sponge::get_ale_sponge_thicknesses(G G, data_h data_h, sponge_mask sponge_mask, CS CS) Return the thicknesses used for the data with a fixed ALE sponge.
- Parameters
[in] g: The ocean’s grid structure (in).[inout] data_h: The thicknesses of the sponge input layers [H ~> m or kg m-2].[out] sponge_mask: A logical mask that is true wherecs: A pointer that is set to point to the control structure for the ALE_sponge module.
-
subroutine
initialize_ale_sponge_varying(Iresttime Iresttime, G G, param_file param_file, CS CS)¶ This subroutine determines the number of points which are within sponges in this computational domain. Only points that have positive values of Iresttime and which mask2dT indicates are ocean points are included in the sponges.
- Parameters
[in] g: The ocean’s grid structure.[in] iresttime: The inverse of the restoring time [s-1].[in] param_file: A structure indicating the open file to parse for model parameter values.cs: A pointer that is set to point to the control structure for this module (in/out).
-
subroutine, public mom_ale_sponge::init_ale_sponge_diags(Time Time, G G, diag diag, CS CS) Initialize diagnostics for the ALE_sponge module.
- Parameters
[in] time: The current model time[in] g: The ocean’s grid structure[inout] diag: A structure that is used to regulate diagnostic output.cs: ALE sponge control structure
-
subroutine
set_up_ale_sponge_field_fixed(sp_val sp_val, G G, f_ptr f_ptr, CS CS)¶ This subroutine stores the reference profile at h points for the variable whose address is given by f_ptr.
- Parameters
[in] g: Grid structurecs: ALE sponge control structure (in/out).[in] sp_val: Field to be used in the sponge, it has arbritary number of layers.[in] f_ptr: Pointer to the field to be damped
-
subroutine
set_up_ale_sponge_field_varying(filename filename, fieldname fieldname, Time Time, G G, GV GV, f_ptr f_ptr, CS CS)¶ This subroutine stores the reference profile at h points for the variable whose address is given by filename and fieldname.
- Parameters
[in] filename: The name of the file with the time varying field data[in] fieldname: The name of the field in the file with the time varying field data[in] time: The current model time[in] g: Grid structure (in).[in] gv: ocean vertical grid structure[in] f_ptr: Pointer to the field to be damped (in).cs: Sponge control structure (in/out).
-
subroutine
set_up_ale_sponge_vel_field_fixed(u_val u_val, v_val v_val, G G, u_ptr u_ptr, v_ptr v_ptr, CS CS)¶ This subroutine stores the reference profile at u and v points for the variable whose address is given by u_ptr and v_ptr.
- Parameters
[in] g: Grid structure (in).cs: Sponge structure (in/out).[in] u_val: u field to be used in the sponge, it has arbritary number of layers.[in] v_val: v field to be used in the sponge, it has arbritary number of layers.[in] u_ptr: u pointer to the field to be damped[in] v_ptr: v pointer to the field to be damped
-
subroutine
set_up_ale_sponge_vel_field_varying(filename_u filename_u, fieldname_u fieldname_u, filename_v filename_v, fieldname_v fieldname_v, Time Time, G G, US US, CS CS, u_ptr u_ptr, v_ptr v_ptr)¶ This subroutine stores the reference profile at uand v points for the variable whose address is given by u_ptr and v_ptr.
- Parameters
[in] filename_u: File name for u field[in] fieldname_u: Name of u variable in file[in] filename_v: File name for v field[in] fieldname_v: Name of v variable in file[in] time: Model time[inout] g: Ocean grid (in)[in] us: A dimensional unit scaling typecs: Sponge structure (in/out).[in] u_ptr: u pointer to the field to be damped (in).[in] v_ptr: v pointer to the field to be damped (in).
-
subroutine, public mom_ale_sponge::apply_ale_sponge(h h, dt dt, G G, GV GV, US US, CS CS, Time Time) This subroutine applies damping to the layers thicknesses, temp, salt and a variety of tracers for every column where there is damping.
- Parameters
[inout] g: The ocean’s grid structure (in).[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] h: Layer thickness [H ~> m or kg m-2] (in)[in] dt: The amount of time covered by this call [s].cs: A pointer to the control structure for this module that is set by a previous call to initialize_sponge (in).[in] time: The current model date
-
subroutine, public mom_ale_sponge::ale_sponge_end(CS CS) This subroutine deallocates any memory associated with the ALE_sponge module.
- Parameters
cs: A pointer to the control structure that is set by a previous call to initialize_sponge.
-
namespace
mom_barotropic¶ Baropotric solver.
By Robert Hallberg, April 1994 - January 2007
This program contains the subroutines that time steps the linearized barotropic equations. btstep is used to actually time step the barotropic equations, and contains most of the substance of this module.
btstep uses a forwards-backwards based scheme to time step the barotropic equations, returning the layers’ accelerations due to the barotropic changes in the ocean state, the final free surface height (or column mass), and the volume (or mass) fluxes summed through the layers and averaged over the baroclinic time step. As input, btstep takes the initial 3-D velocities, the inital free surface height, the 3-D accelerations of the layers, and the external forcing. Everything in btstep is cast in terms of anomalies, so if everything is in balance, there is explicitly no acceleration due to btstep.
The spatial discretization of the continuity equation is second order accurate. A flux conservative form is used to guarantee global conservation of volume. The spatial discretization of the momentum equation is second order accurate. The Coriolis force is written in a form which does not contribute to the energy tendency and which conserves linearized potential vorticity, f/D. These terms are exactly removed from the baroclinic momentum equations, so the linearization of vorticity advection will not degrade the overall solution.
btcalc calculates the fractional thickness of each layer at the velocity points, for later use in calculating the barotropic velocities and the averaged accelerations. Harmonic mean thicknesses (i.e. 2*h_L*h_R/(h_L + h_R)) are used to avoid overly strong weighting of overly thin layers. This may later be relaxed to use thicknesses determined from the continuity equations.
bt_mass_source determines the real mass sources for the barotropic solver, along with the corrective pseudo-fluxes that keep the barotropic and baroclinic estimates of the free surface height close to each other. Given the layer thicknesses and the free surface height that correspond to each other, it calculates a corrective mass source that is added to the barotropic continuity* equation, and optionally adjusts a slowly varying correction rate. Newer algorithmic changes have deemphasized the need for this, but it is still here to add net water sources to the barotropic solver.*
barotropic_init allocates and initializes any barotropic arrays that have not been read from a restart file, reads parameters from the inputfile, and sets up diagnostic fields.
barotropic_end deallocates anything allocated in barotropic_init or register_barotropic_restarts.
register_barotropic_restarts is used to indicate any fields that are private to the barotropic solver that need to be included in the restart files, and to ensure that they are read.
Unnamed Group
-
integer
id_clock_sync= -1¶ CPU time clock IDs.
-
integer
id_clock_calc= -1¶ CPU time clock IDs.
-
integer
id_clock_calc_pre= -1¶ CPU time clock IDs.
-
integer
id_clock_calc_post= -1¶ CPU time clock IDs.
-
integer
id_clock_pass_step= -1¶ CPU time clock IDs.
-
integer
id_clock_pass_pre= -1¶ CPU time clock IDs.
-
integer
id_clock_pass_post= -1¶ CPU time clock IDs.
-
integer, parameter mom_barotropic::harmonic = 1 Enumeration values for various schemes.
-
integer, parameter mom_barotropic::arithmetic = 2 CPU time clock IDs.
-
integer, parameter mom_barotropic::hybrid = 3 CPU time clock IDs.
-
integer, parameter mom_barotropic::from_bt_cont = 4 CPU time clock IDs.
-
integer, parameter mom_barotropic::hybrid_bt_cont = 5 CPU time clock IDs.
-
character*(20), parameter mom_barotropic::hybrid_string = "HYBRID" CPU time clock IDs.
-
character*(20), parameter mom_barotropic::harmonic_string = "HARMONIC" CPU time clock IDs.
-
character*(20), parameter mom_barotropic::arithmetic_string = "ARITHMETIC" CPU time clock IDs.
-
character*(20), parameter mom_barotropic::bt_cont_string = "FROM_BT_CONT" CPU time clock IDs.
-
subroutine, public mom_barotropic::btstep(U_in U_in, V_in V_in, eta_in eta_in, dt dt, bc_accel_u bc_accel_u, bc_accel_v bc_accel_v, forces forces, pbce pbce, eta_PF_in eta_PF_in, U_Cor U_Cor, V_Cor V_Cor, accel_layer_u accel_layer_u, accel_layer_v accel_layer_v, eta_out eta_out, uhbtav uhbtav, vhbtav vhbtav, G G, GV GV, US US, CS CS, visc_rem_u visc_rem_u, visc_rem_v visc_rem_v, etaav etaav, OBC OBC, BT_cont BT_cont, eta_PF_start eta_PF_start, taux_bot taux_bot, tauy_bot tauy_bot, uh0 uh0, vh0 vh0, u_uh0 u_uh0, v_vh0 v_vh0) This subroutine time steps the barotropic equations explicitly. For gravity waves, anything between a forwards-backwards scheme and a simulated backwards Euler scheme is used, with bebt between 0.0 and 1.0 determining the scheme. In practice, bebt must be of order 0.2 or greater. A forwards-backwards treatment of the Coriolis terms is always used.
- Parameters
[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] u_in: The initial (3-D) zonal velocity [m s-1].[in] v_in: The initial (3-D) meridional velocity [m s-1].[in] eta_in: The initial barotropic free surface height anomaly or column mass anomaly [H ~> m or kg m-2].[in] dt: The time increment to integrate over.[in] bc_accel_u: The zonal baroclinic accelerations [m s-2].[in] bc_accel_v: The meridional baroclinic accelerations, [m s-2].[in] forces: A structure with the driving mechanical forces[in] pbce: The baroclinic pressure anomaly in each layer due to free surface height anomalies [L2 H-1 T-2 ~> m s-2 or m4 kg-1 s-2].[in] eta_pf_in: The 2-D eta field (either SSH anomaly or column mass anomaly) that was used to calculate the input pressure gradient accelerations (or its final value if eta_PF_start is provided [H ~> m or kg m-2]. Note: eta_in, pbce, and eta_PF_in must have up-to-date values in the first point of their halos.[in] u_cor: The (3-D) zonal-velocities used to calculate the Coriolis terms in bc_accel_u [m s-1].[in] v_cor: Ditto for meridonal bc_accel_v.[out] accel_layer_u: The zonal acceleration of each layer due to the barotropic calculation [m s-2].[out] accel_layer_v: The meridional acceleration of each layer due to the barotropic calculation [m s-2].[out] eta_out: The final barotropic free surface height anomaly or column mass anomaly [H ~> m or kg m-2].[out] uhbtav: the barotropic zonal volume or mass fluxes averaged through the barotropic steps [H m2 s-1 ~> m3 or kg s-1].[out] vhbtav: the barotropic meridional volume or mass fluxes averaged through the barotropic steps [H m2 s-1 ~> m3 or kg s-1].cs: The control structure returned by a previous call to barotropic_init.[in] visc_rem_u: Both the fraction of the momentum originally in a layer that remains after a time-step of viscosity, and the fraction of a time-step’s worth of a barotropic acceleration that a layer experiences after viscosity is applied, in the zonal direction. Nondimensional between 0 (at the bottom) and 1 (far above the bottom).[in] visc_rem_v: Ditto for meridional direction [nondim].[out] etaav: The free surface height or column mass averaged over the barotropic integration [H ~> m or kg m-2].obc: The open boundary condition structure.bt_cont: A structure with elements that describe the effective open face areas as a function of barotropic flow.eta_pf_start: The eta field consistent with the pressure gradient at the start of the barotropic stepping [H ~> m or kg m-2].taux_bot: The zonal bottom frictional stress from ocean to the seafloor [Pa].tauy_bot: The meridional bottom frictional stress from ocean to the seafloor [Pa].uh0: The zonal layer transports at reference velocities [H m s-1 ~> m2 s-1 or kg m-1 s-1].u_uh0: The velocities used to calculate uh0 [m s-1]vh0: The zonal layer transports at reference velocities [H m s-1 ~> m2 s-1 or kg m-1 s-1].v_vh0: The velocities used to calculate vh0 [m s-1]
-
subroutine, public mom_barotropic::set_dtbt(G G, GV GV, US US, CS CS, eta eta, pbce pbce, BT_cont BT_cont, gtot_est gtot_est, SSH_add SSH_add) This subroutine automatically determines an optimal value for dtbt based on some state of the ocean.
- Parameters
[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling typecs: Barotropic control structure.[in] eta: The barotropic free surface height anomaly or column mass anomaly [H ~> m or kg m-2].[in] pbce: The baroclinic pressure anomaly in each layer due to free surface height anomalies [L2 H-1 T-2 ~> m s-2 or m4 kg-1 s-2].bt_cont: A structure with elements that describe the effective open face areas as a function of barotropic flow.[in] gtot_est: An estimate of the total gravitational acceleration [L2 Z-1 T-2 ~> m s-2].[in] ssh_add: An additional contribution to SSH to provide a margin of error when calculating the external wave speed [Z ~> m].
-
subroutine
apply_velocity_obcs(OBC OBC, ubt ubt, vbt vbt, uhbt uhbt, vhbt vhbt, ubt_trans ubt_trans, vbt_trans vbt_trans, eta eta, ubt_old ubt_old, vbt_old vbt_old, BT_OBC BT_OBC, G G, MS MS, US US, halo halo, dtbt dtbt, bebt bebt, use_BT_cont use_BT_cont, Datu Datu, Datv Datv, BTCL_u BTCL_u, BTCL_v BTCL_v, uhbt0 uhbt0, vhbt0 vhbt0)¶ The following 4 subroutines apply the open boundary conditions. This subroutine applies the open boundary conditions on barotropic velocities and mass transports, as developed by Mehmet Ilicak.
- Parameters
obc: An associated pointer to an OBC type.[inout] g: The ocean’s grid structure.[in] ms: A type that describes the memory sizes of the argument arrays.[inout] ubt: the zonal barotropic velocity [m s-1].[inout] uhbt: the zonal barotropic transport [H m2 s-1 ~> m3 s-1 or kg s-1].[inout] ubt_trans: the zonal barotropic velocity used in transport [L T-1 ~> m s-1].[inout] vbt: the meridional barotropic velocity [m s-1].[inout] vhbt: the meridional barotropic transport [H m2 s-1 ~> m3 s-1 or kg s-1].[inout] vbt_trans: the meridional BT velocity used in transports [m s-1].[in] eta: The barotropic free surface height anomaly or column mass anomaly [H ~> m or kg m-2].[in] ubt_old: The starting value of ubt in a barotropic step [L T-1 ~> m s-1].[in] vbt_old: The starting value of vbt in a barotropic step [L T-1 ~> m s-1].[in] bt_obc: A structure with the private barotropic arrays related to the open boundary conditions, set by set_up_BT_OBC.[in] us: A dimensional unit scaling type[in] halo: The extra halo size to use here.[in] dtbt: The time step [T ~> s].[in] bebt: The fractional weighting of the future velocity in determining the transport.[in] use_bt_cont: If true, use the BT_cont_types to calculate transports.[in] datu: A fixed estimate of the face areas at u points [H L ~> m2 or kg m-1].[in] datv: A fixed estimate of the face areas at v points [H L ~> m2 or kg m-1].[in] btcl_u: Structure of information used for a dynamic estimate of the face areas at u-points.[in] btcl_v: Structure of information used for a dynamic estimate of the face areas at v-points.[in] uhbt0: A correction to the zonal transport so that the barotropic functions agree with the sum of the layer transports [H L2 T-1 ~> m3 s-1 or kg s-1].[in] vhbt0: A correction to the meridional transport so that the barotropic functions agree with the sum of the layer transports [H L2 T-1 ~> m3 s-1 or kg s-1].
-
subroutine
set_up_bt_obc(OBC OBC, eta eta, BT_OBC BT_OBC, BT_Domain BT_Domain, G G, GV GV, US US, MS MS, halo halo, use_BT_cont use_BT_cont, Datu Datu, Datv Datv, BTCL_u BTCL_u, BTCL_v BTCL_v)¶ This subroutine sets up the private structure used to apply the open boundary conditions, as developed by Mehmet Ilicak.
- Parameters
obc: An associated pointer to an OBC type.[in] ms: A type that describes the memory sizes of the argument arrays.[in] eta: The barotropic free surface height anomaly or column mass anomaly [H ~> m or kg m-2].[inout] bt_obc: A structure with the private barotropic arrays related to the open boundary conditions, set by set_up_BT_OBC.[inout] bt_domain: MOM_domain_type associated with wide arrays[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] halo: The extra halo size to use here.[in] use_bt_cont: If true, use the BT_cont_types to calculate transports.[in] datu: A fixed estimate of the face areas at u points [L m ~> m2 or kg m-1].[in] datv: A fixed estimate of the face areas at v points [L m ~> m2 or kg m-1].[in] btcl_u: Structure of information used for a dynamic estimate of the face areas at u-points.[in] btcl_v: Structure of information used for a dynamic estimate of the face areas at v-points.
-
subroutine
destroy_bt_obc(BT_OBC BT_OBC)¶ Clean up the BT_OBC memory.
- Parameters
[inout] bt_obc: A structure with the private barotropic arrays related to the open boundary conditions, set by set_up_BT_OBC.
-
subroutine, public mom_barotropic::btcalc(h h, G G, GV GV, CS CS, h_u h_u, h_v h_v, may_use_default may_use_default, OBC OBC) btcalc calculates the barotropic velocities from the full velocity and thickness fields, determines the fraction of the total water column in each layer at velocity points, and determines a corrective fictitious mass source that will drive the barotropic estimate of the free surface height toward the baroclinic estimate.
- Parameters
[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] h: Layer thicknesses [H ~> m or kg m-2].cs: The control structure returned by a previous call to barotropic_init.[in] h_u: The specified thicknesses at u-points [H ~> m or kg m-2].[in] h_v: The specified thicknesses at v-points [H ~> m or kg m-2].[in] may_use_default: An optional logical argument to indicate that the default velocity point thicknesses may be used for this particular calculation, even though the setting of CShvel_scheme would usually require that h_u and h_v be passed in.obc: Open boundary control structure.
-
real function mom_barotropic::find_uhbt(u u, BTC BTC, US US) The function find_uhbt determines the zonal transport for a given velocity.
- Return
The zonal barotropic transport [L2 H T-1 ~> m3 s-1]
- Parameters
[in] u: The local zonal velocity [L T-1 ~> m s-1][in] btc: A structure containing various fields that allow the barotropic transports to be calculated consistently with the layers’ continuity equations.[in] us: A dimensional unit scaling type
-
real function mom_barotropic::uhbt_to_ubt(uhbt uhbt, BTC BTC, US US, guess guess) This function inverts the transport function to determine the barotopic velocity that is consistent with a given transport.
- Return
The result - The velocity that gives uhbt transport [m s-1].
- Parameters
[in] uhbt: The barotropic zonal transport that should be inverted for, [H L2 T-1 ~> m3 s-1 or kg s-1].[in] btc: A structure containing various fields that allow the barotropic transports to be calculated consistently with the layers’ continuity equations.[in] us: A dimensional unit scaling type[in] guess: A guess at what ubt will be [L T-1 ~> m s-1]. The result is not allowed to be dramatically larger than guess.
-
real function mom_barotropic::find_vhbt(v v, BTC BTC, US US) The function find_vhbt determines the meridional transport for a given velocity.
- Return
The meridional barotropic transport [L2 H T-1 ~> m3 s-1]
- Parameters
[in] v: The local meridional velocity [L T-1 ~> m s-1][in] btc: A structure containing various fields that allow the barotropic transports to be calculated consistently with the layers’ continuity equations.[in] us: A dimensional unit scaling type
-
real function mom_barotropic::vhbt_to_vbt(vhbt vhbt, BTC BTC, US US, guess guess) This function inverts the transport function to determine the barotopic velocity that is consistent with a given transport.
- Return
The result - The velocity that gives vhbt transport [L T-1 ~> m s-1].
- Parameters
[in] vhbt: The barotropic meridional transport that should be inverted for [H L2 T-1 ~> m3 s-1 or kg s-1].[in] btc: A structure containing various fields that allow the barotropic transports to be calculated consistently with the layers’ continuity equations.[in] us: A dimensional unit scaling type[in] guess: A guess at what vbt will be. The result is not allowed to be dramatically larger than guess [L T-1 ~> m s-1].
-
subroutine
set_local_bt_cont_types(BT_cont BT_cont, BTCL_u BTCL_u, BTCL_v BTCL_v, G G, US US, MS MS, BT_Domain BT_Domain, halo halo)¶ This subroutine sets up reordered versions of the BT_cont type in the local_BT_cont types, which have wide halos properly filled in.
- Parameters
[inout] bt_cont: The BT_cont_type input to the barotropic solver.[in] ms: A type that describes the memory sizes of the argument arrays.[out] btcl_u: A structure with the u information from BT_cont.[out] btcl_v: A structure with the v information from BT_cont.[in] g: The ocean’s grid structure.[in] us: A dimensional unit scaling type[inout] bt_domain: The domain to use for updating the halos of wide arrays.[in] halo: The extra halo size to use here.
-
subroutine
adjust_local_bt_cont_types(ubt ubt, uhbt uhbt, vbt vbt, vhbt vhbt, BTCL_u BTCL_u, BTCL_v BTCL_v, G G, US US, MS MS, halo halo)¶ Adjust_local_BT_cont_types sets up reordered versions of the BT_cont type in the local_BT_cont types, which have wide halos properly filled in.
- Parameters
[in] ms: A type that describes the memory sizes of the argument arrays.[in] ubt: The linearization zonal barotropic velocity [m s-1].[in] uhbt: The linearization zonal barotropic transport[in] vbt: The linearization meridional barotropic velocity [m s-1].[in] vhbt: The linearization meridional barotropic transport[out] btcl_u: A structure with the u information from BT_cont.[out] btcl_v: A structure with the v information from BT_cont.[in] g: The ocean’s grid structure.[in] us: A dimensional unit scaling type[in] halo: The extra halo size to use here.
-
subroutine
bt_cont_to_face_areas(BT_cont BT_cont, Datu Datu, Datv Datv, G G, US US, MS MS, halo halo, maximize maximize)¶ This subroutine uses the BTCL types to find typical or maximum face areas, which can then be used for finding wave speeds, etc.
- Parameters
[inout] bt_cont: The BT_cont_type input to the barotropic solver.[in] ms: A type that describes the memory sizes of the argument arrays.[out] datu: The effective zonal face area [H L ~> m2 or kg m-1].[out] datv: The effective meridional face area [H L ~> m2 or kg m-1].[in] g: The ocean’s grid structure.[in] us: A dimensional unit scaling type[in] halo: The extra halo size to use here.[in] maximize: If present and true, find the maximum face area for any velocity.
-
subroutine
swap(a a, b b)¶ Swap the values of two real variables.
- Parameters
[inout] a: The first variable to be swapped.[inout] b: The second variable to be swapped.
-
subroutine
find_face_areas(Datu Datu, Datv Datv, G G, GV GV, US US, CS CS, MS MS, eta eta, halo halo, add_max add_max)¶ This subroutine determines the open face areas of cells for calculating the barotropic transport.
- Parameters
[in] ms: A type that describes the memory sizes of the argument arrays.[out] datu: The open zonal face area [H L ~> m2 or kg m-1].[out] datv: The open meridional face area [H L ~> m2 or kg m-1].[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling typecs: The control structure returned by a previous call to barotropic_init.[in] eta: The barotropic free surface height anomaly[in] halo: The halo size to use, default = 1.[in] add_max: A value to add to the maximum depth (used to overestimate the external wave speed) [Z ~> m].
-
subroutine, public mom_barotropic::bt_mass_source(h h, eta eta, set_cor set_cor, G G, GV GV, CS CS) bt_mass_source determines the appropriately limited mass source for the barotropic solver, along with a corrective fictitious mass source that will drive the barotropic estimate of the free surface height toward the baroclinic estimate.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] h: Layer thicknesses [H ~> m or kg m-2].[in] eta: The free surface height that is to be corrected [H ~> m or kg m-2].[in] set_cor: A flag to indicate whether to set the corrective fluxes (and update the slowly varying part of eta_cor) (.true.) or whether to incrementally update the corrective fluxes.cs: The control structure returned by a previous call to barotropic_init.
-
subroutine, public mom_barotropic::barotropic_init(u u, v v, h h, eta eta, Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS, restart_CS restart_CS, calc_dtbt calc_dtbt, BT_cont BT_cont, tides_CSp tides_CSp) barotropic_init initializes a number of time-invariant fields used in the barotropic calculation and initializes any barotropic fields that have not already been initialized.
- Parameters
[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] u: The zonal velocity [m s-1].[in] v: The meridional velocity [m s-1].[in] h: Layer thicknesses [H ~> m or kg m-2].[in] eta: Free surface height or column mass anomaly[in] time: The current model time.[in] param_file: A structure to parse for run-time parameters.[inout] diag: A structure that is used to regulate diagnostic output.cs: A pointer to the control structure for this module that is set in register_barotropic_restarts.restart_cs: A pointer to the restart control structure.[out] calc_dtbt: If true, the barotropic time step must be recalculated before stepping.bt_cont: A structure with elements that describe thetides_csp: A pointer to the control structure of the
-
subroutine, public mom_barotropic::barotropic_get_tav(CS CS, ubtav ubtav, vbtav vbtav, G G, US US) Copies ubtav and vbtav from private type into arrays.
- Parameters
cs: Control structure for this module[in] g: Grid structure[inout] ubtav: Zonal barotropic velocity averaged over a baroclinic timestep [m s-1][inout] vbtav: Meridional barotropic velocity averaged over a baroclinic timestep [m s-1][in] us: A dimensional unit scaling type
-
subroutine, public mom_barotropic::barotropic_end(CS CS) Clean up the barotropic control structure.
- Parameters
cs: Control structure to clear out.
-
subroutine, public mom_barotropic::register_barotropic_restarts(HI HI, GV GV, param_file param_file, CS CS, restart_CS restart_CS) This subroutine is used to register any fields from MOM_barotropic.F90 that should be written to or read from the restart file.
- Parameters
[in] hi: A horizontal index type structure.[in] param_file: A structure to parse for run-time parameters.cs: A pointer that is set to point to the control structure for this module.[in] gv: The ocean’s vertical grid structure.restart_cs: A pointer to the restart control structure.
-
integer
-
namespace
mom_bkgnd_mixing¶ Interface to background mixing schemes, including the Bryan and Lewis (1979) which is applied via CVMix.
Functions
-
subroutine, public mom_bkgnd_mixing::bkgnd_mixing_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS) Initialize the background mixing routine.
- Parameters
[in] time: The current time.[in] g: Grid structure.[in] gv: Vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: Run-time parameter file handle[inout] diag: Diagnostics control structure.cs: This module’s control structure.
-
subroutine, public mom_bkgnd_mixing::sfc_bkgnd_mixing(G G, US US, CS CS) Get surface vertical background diffusivities/viscosities.
- Parameters
[in] g: Grid structure.[in] us: A dimensional unit scaling type[inout] cs: The control structure returned by a previous call to bkgnd_mixing_init.
-
subroutine, public mom_bkgnd_mixing::calculate_bkgnd_mixing(h h, tv tv, N2_lay N2_lay, Kd_lay Kd_lay, Kv Kv, j j, G G, GV GV, US US, CS CS) Calculates the vertical background diffusivities/viscosities.
- Parameters
[in] g: Grid structure.[in] gv: Vertical grid structure.[in] us: A dimensional unit scaling type[in] h: Layer thickness [H ~> m or kg m-2].[in] tv: Thermodynamics structure.[in] n2_lay: squared buoyancy frequency associated with layers [T-2 ~> s-2][inout] kd_lay: Diapycnal diffusivity of each layer [Z2 T-1 ~> m2 s-1].kv: The “slow” vertical viscosity at each interface (not layer!) [Z2 T-1 ~> m2 s-1][in] j: Meridional grid indexcs: The control structure returned by a previous call to bkgnd_mixing_init.
-
logical function mom_bkgnd_mixing::cvmix_bkgnd_is_used(param_file param_file) Reads the parameter “USE_CVMix_BACKGROUND” and returns state. This function allows other modules to know whether this parameterization will be used without needing to duplicate the log entry.
- Parameters
[in] param_file: A structure to parse for run-time parameters
-
subroutine
check_bkgnd_scheme(CS CS, str str)¶ Sets CSbkgnd_scheme_str to check whether multiple background diffusivity schemes are activated. The string is also for error/log messages.
- Parameters
cs: Control structure[in] str: Background scheme identifier deducted from MOM_input parameters
-
subroutine, public mom_bkgnd_mixing::bkgnd_mixing_end(CS CS) Clear pointers and dealocate memory.
- Parameters
cs: Control structure for this module that will be deallocated in this subroutine
Variables
-
character(len=40) mom_bkgnd_mixing::mdl = "MOM_bkgnd_mixing" This module’s name.
-
-
namespace
mom_boundary_update¶ Controls where open boundary conditions are applied.
This module updates the open boundary arrays when time-varying. It caused a circular dependency with the tidal_bay setup when MOM_open_boundary.
A small fragment of the grid is shown below:
j+1 x ^ x ^ x At x: q, CoriolisBu j+1 > o > o > At ^: v, tauy j x ^ x ^ x At >: u, taux j > o > o > At o: h, bathyT, buoy, tr, T, S, Rml, ustar j-1 x ^ x ^ x i-1 i i+1 At x & ^: i i+1 At > & o:
The boundaries always run through q grid points (x).
Unnamed Group
-
integer
id_clock_pass¶ A CPU time clock ID.
-
subroutine, public mom_boundary_update::call_obc_register(param_file param_file, CS CS, OBC OBC) The following subroutines and associated definitions provide the machinery to register and call the subroutines that initialize open boundary conditions.
- Parameters
[in] param_file: Parameter file to parsecs: Control structure for OBCsobc: Open boundary structure
-
subroutine, public mom_boundary_update::update_obc_data(OBC OBC, G G, GV GV, US US, tv tv, h h, CS CS, Time Time) Calls appropriate routine to update the open boundary conditions.
- Parameters
[in] g: Ocean grid structure[in] gv: Ocean vertical grid structure[in] us: A dimensional unit scaling type[in] tv: Thermodynamics structure[inout] h: layer thicknesses [H ~> m or kg m-2]obc: Open boundary structurecs: Control structure for OBCs[in] time: Model time
-
subroutine, public mom_boundary_update::obc_register_end(CS CS) Clean up the OBC registry.
- Parameters
cs: Control structure for OBCs
-
integer
-
namespace
mom_bulk_mixed_layer¶ Build mixed layer parameterization.
By Robert Hallberg, 1997 - 2005.
This file contains the subroutine (bulkmixedlayer) that implements a Kraus-Turner-like bulk mixed layer, based on the work of various people, as described in the review paper by Niiler and Kraus (1979), with particular attention to the form proposed by Oberhuber (JPO, 1993, 808-829), with an extension to a refied bulk mixed layer as described in Hallberg (Aha Huliko’a, 2003). The physical processes portrayed in this subroutine include convective adjustment and mixed layer entrainment and detrainment. Penetrating shortwave radiation and an exponential decay of TKE fluxes are also supported by this subroutine. Several constants can alternately be set to give a traditional Kraus-Turner mixed layer scheme, although that is not the preferred option. The physical processes and arguments are described in detail below.
Unnamed Group
-
integer
id_clock_detrain= 0¶ CPU clock IDs.
-
integer
id_clock_mech= 0¶ CPU clock IDs.
-
integer
id_clock_conv= 0¶ CPU clock IDs.
-
integer
id_clock_adjustment= 0¶ CPU clock IDs.
-
integer
id_clock_eos= 0¶ CPU clock IDs.
-
integer
id_clock_resort= 0¶ CPU clock IDs.
-
integer
id_clock_pass= 0¶ CPU clock IDs.
-
subroutine, public mom_bulk_mixed_layer::bulkmixedlayer(h_3d h_3d, u_3d u_3d, v_3d v_3d, tv tv, fluxes fluxes, dt_in_T dt_in_T, ea ea, eb eb, G G, GV GV, US US, CS CS, optics optics, Hml Hml, aggregate_FW_forcing aggregate_FW_forcing, dt_diag dt_diag, last_call last_call) This subroutine partially steps the bulk mixed layer model. The following processes are executed, in the order listed.
Undergo convective adjustment into mixed layer.
Apply surface heating and cooling.
Starting from the top, entrain whatever fluid the TKE budget permits. Penetrating shortwave radiation is also applied at this point.
If there is any unentrained fluid that was formerly in the mixed layer, detrain this fluid into the buffer layer. This is equivalent to the mixed layer detraining to the Monin- Obukhov depth.
Divide the fluid in the mixed layer evenly into CSnkml pieces.
Split the buffer layer if appropriate. Layers 1 to nkml are the mixed layer, nkml+1 to nkml+nkbl are the buffer layers. The results of this subroutine are mathematically identical if there are multiple pieces of the mixed layer with the same density or if there is just a single layer. There is no stability limit on the time step.
The key parameters for the mixed layer are found in the control structure. These include mstar, nstar, nstar2, pen_SW_frac, pen_SW_scale, and TKE_decay. For the Oberhuber (1993) mixed layer, the values of these are: pen_SW_frac = 0.42, pen_SW_scale = 15.0 m, mstar = 1.25, nstar = 1, TKE_decay = 2.5, conv_decay = 0.5 TKE_decay is 1/kappa in eq. 28 of Oberhuber (1993), while conv_decay is 1/mu. Conv_decay has been eliminated in favor of the well-calibrated form for the efficiency of penetrating convection from Wang (2003). For a traditional Kraus-Turner mixed layer, the values are: pen_SW_frac = 0.0, pen_SW_scale = 0.0 m, mstar = 1.25, nstar = 0.4, TKE_decay = 0.0, conv_decay = 0.0
- Parameters
[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[inout] h_3d: Layer thickness [H ~> m or kg m-2].[in] u_3d: Zonal velocities interpolated to h points[in] v_3d: Zonal velocities interpolated to h points[inout] tv: A structure containing pointers to any available thermodynamic fields. Absent fields have NULL ptrs.[inout] fluxes: A structure containing pointers to any possible forcing fields. Unused fields have NULL ptrs.[in] dt_in_t: Time increment [T ~> s].[inout] ea: The amount of fluid moved downward into a[inout] eb: The amount of fluid moved upward into acs: The control structure returned by a previous call to mixedlayer_init.optics: The structure containing the inverse of the vertical absorption decay scale for penetrating shortwave radiation [m-1].hml: Active mixed layer depth [m].[in] aggregate_fw_forcing: If true, the net incoming and outgoing surface freshwater fluxes are combined before being applied, instead of being applied separately.[in] dt_diag: The diagnostic time step, which may be less than dt if there are two callse to mixedlayer [T ~> s].[in] last_call: if true, this is the last call to mixedlayer in the current time step, so diagnostics will be written. The default is .true.
-
subroutine
convective_adjustment(h h, u u, v v, R0 R0, Rcv Rcv, T T, S S, eps eps, d_eb d_eb, dKE_CA dKE_CA, cTKE cTKE, j j, G G, GV GV, US US, CS CS, nz_conv nz_conv)¶ This subroutine does instantaneous convective entrainment into the buffer layers and mixed layers to remove hydrostatic instabilities. Any water that is lighter than currently in the mixed- or buffer- layer is entrained.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[inout] h: Layer thickness [H ~> m or kg m-2]. The units of h are referred to as H below.[inout] u: Zonal velocities interpolated to h points, m s-1.[inout] v: Zonal velocities interpolated to h points, m s-1.[inout] t: Layer temperatures [degC].[inout] s: Layer salinities [ppt].[inout] r0: Potential density referenced to surface pressure [kg m-3].[inout] rcv: The coordinate defining potential density [kg m-3].[inout] d_eb: The downward increase across a layer in the entrainment from below [H ~> m or kg m-2]. Positive values go with mass gain by a layer.[in] eps: The negligibly small amount of water that will be left in each layer [H ~> m or kg m-2].[out] dke_ca: The vertically integrated change in kinetic energy due to convective adjustment [Z m2 T-2 ~> m3 s-2].[out] ctke: The buoyant turbulent kinetic energy source due to convective adjustment [Z m2 T-2 ~> m3 s-2].[in] j: The j-index to work on.[in] us: A dimensional unit scaling typecs: The control structure for this module.[in] nz_conv: If present, the number of layers over which to do convective adjustment (perhaps CSnkml).
-
subroutine
mixedlayer_convection(h h, d_eb d_eb, htot htot, Ttot Ttot, Stot Stot, uhtot uhtot, vhtot vhtot, R0_tot R0_tot, Rcv_tot Rcv_tot, u u, v v, T T, S S, R0 R0, Rcv Rcv, eps eps, dR0_dT dR0_dT, dRcv_dT dRcv_dT, dR0_dS dR0_dS, dRcv_dS dRcv_dS, netMassInOut netMassInOut, netMassOut netMassOut, Net_heat Net_heat, Net_salt Net_salt, nsw nsw, Pen_SW_bnd Pen_SW_bnd, opacity_band opacity_band, Conv_en Conv_en, dKE_FC dKE_FC, j j, ksort ksort, G G, GV GV, US US, CS CS, tv tv, fluxes fluxes, dt_in_T dt_in_T, aggregate_FW_forcing aggregate_FW_forcing)¶ This subroutine causes the mixed layer to entrain to the depth of free convection. The depth of free convection is the shallowest depth at which the fluid is denser than the average of the fluid above.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[inout] h: Layer thickness [H ~> m or kg m-2].[inout] d_eb: The downward increase across a layer in the[out] htot: The accumulated mixed layer thickness [H ~> m or kg m-2].[out] ttot: The depth integrated mixed layer temperature [degC H ~> degC m or degC kg m-2].[out] stot: The depth integrated mixed layer salinity [ppt H ~> ppt m or ppt kg m-2].[out] uhtot: The depth integrated mixed layer zonal velocity, H m s-1.[out] vhtot: The integrated mixed layer meridional velocity, H m s-1.[out] r0_tot: The integrated mixed layer potential density referenced to 0 pressure [H kg m-2 ~> kg m-1 or kg2 m-4].[out] rcv_tot: The integrated mixed layer coordinate variable potential density [H kg m-2 ~> kg m-1 or kg2 m-4].[in] u: Zonal velocities interpolated to h points, m s-1.[in] v: Zonal velocities interpolated to h points, m s-1.[in] t: Layer temperatures [degC].[in] s: Layer salinities [ppt].[in] r0: Potential density referenced to[in] rcv: The coordinate defining potential[in] eps: The negligibly small amount of water[in] dr0_dt: The partial derivative of R0 with respect to temperature [kg m-3 degC-1].[in] drcv_dt: The partial derivative of Rcv with respect to temperature [kg m-3 degC-1].[in] dr0_ds: The partial derivative of R0 with respect to salinity [kg m-3 ppt-1].[in] drcv_ds: The partial derivative of Rcv with respect to salinity [kg m-3 ppt-1].[in] netmassinout: The net mass flux (if non-Boussinesq) or volume flux (if Boussinesq) into the ocean within a time step [H ~> m or kg m-2]. (I.e. P+R-E.)[in] netmassout: The mass or volume flux out of the ocean within a time step [H ~> m or kg m-2].[in] net_heat: The net heating at the surface over a time step [degC H ~> degC m or degC kg m-2]. Any penetrating shortwave radiation is not included in Net_heat.[in] net_salt: The net surface salt flux into the ocean over a time step [ppt H ~> ppt m or ppt kg m-2].[in] nsw: The number of bands of penetrating shortwave radiation.[inout] pen_sw_bnd: The penetrating shortwave heating at the sea surface in each penetrating band [degC H ~> degC m or degC kg m-2].[in] opacity_band: The opacity in each band of penetrating shortwave radiation [H-1 ~> m-1 or m2 kg-1].[out] conv_en: The buoyant turbulent kinetic energy source due to free convection [Z m2 T-2 ~> m3 s-2].[out] dke_fc: The vertically integrated change in kinetic energy due to free convection [Z m2 T-2 ~> m3 s-2].[in] j: The j-index to work on.[in] ksort: The density-sorted k-indices.[in] us: A dimensional unit scaling typecs: The control structure for this module.[inout] tv: A structure containing pointers to any available thermodynamic fields. Absent fields have NULL ptrs.[inout] fluxes: A structure containing pointers to any possible forcing fields. Unused fields have NULL ptrs.[in] dt_in_t: Time increment [T ~> s].[in] aggregate_fw_forcing: If true, the net incoming and outgoing surface freshwater fluxes are combined before being applied, instead of being applied separately.
-
subroutine
find_starting_tke(htot htot, h_CA h_CA, fluxes fluxes, Conv_En Conv_En, cTKE cTKE, dKE_FC dKE_FC, dKE_CA dKE_CA, TKE TKE, TKE_river TKE_river, Idecay_len_TKE Idecay_len_TKE, cMKE cMKE, dt_in_T dt_in_T, Idt_diag Idt_diag, j j, ksort ksort, G G, GV GV, US US, CS CS)¶ This subroutine determines the TKE available at the depth of free convection to drive mechanical entrainment.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] htot: The accumulated mixed layer thickness [H ~> m or kg m-2][in] h_ca: The mixed layer depth after convective adjustment [H ~> m or kg m-2].[in] fluxes: A structure containing pointers to any possible forcing fields. Unused fields have NULL ptrs.[inout] conv_en: The buoyant turbulent kinetic energy source due to free convection [Z m2 T-2 ~> m3 s-2].[in] dke_fc: The vertically integrated change in kinetic energy due to free convection [Z m2 T-2 ~> m3 s-2].[in] ctke: The buoyant turbulent kinetic energy[in] dke_ca: The vertically integrated change in[out] tke: The turbulent kinetic energy available for mixing over a time step [Z m2 T-2 ~> m3 s-2].[out] idecay_len_tke: The inverse of the vertical decay scale for TKE [H-1 ~> m-1 or m2 kg-1].[in] tke_river: The source of turbulent kinetic energy available for driving mixing at river mouths [Z m2 T-3 ~> m3 s-3].[out] cmke: Coefficients of HpE and HpE^2 in calculating the denominator of MKE_rate, [H-1 ~> m-1 or m2 kg-1] and [H-2 ~> m-2 or m4 kg-2].[in] dt_in_t: The time step [T ~> s].[in] idt_diag: The inverse of the accumulated diagnostic time interval [T-1 ~> s-1].[in] j: The j-index to work on.[in] ksort: The density-sorted k-indicies.cs: The control structure for this module.
-
subroutine
mechanical_entrainment(h h, d_eb d_eb, htot htot, Ttot Ttot, Stot Stot, uhtot uhtot, vhtot vhtot, R0_tot R0_tot, Rcv_tot Rcv_tot, u u, v v, T T, S S, R0 R0, Rcv Rcv, eps eps, dR0_dT dR0_dT, dRcv_dT dRcv_dT, cMKE cMKE, Idt_diag Idt_diag, nsw nsw, Pen_SW_bnd Pen_SW_bnd, opacity_band opacity_band, TKE TKE, Idecay_len_TKE Idecay_len_TKE, j j, ksort ksort, G G, GV GV, US US, CS CS)¶ This subroutine calculates mechanically driven entrainment.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[inout] h: Layer thickness [H ~> m or kg m-2].[inout] d_eb: The downward increase across a layer in the[inout] htot: The accumlated mixed layer thickness [H ~> m or kg m-2].[inout] ttot: The depth integrated mixed layer temperature [degC H ~> degC m or degC kg m-2].[inout] stot: The depth integrated mixed layer salinity [ppt H ~> ppt m or ppt kg m-2].[inout] uhtot: The depth integrated mixed layer zonal velocity, H m s-1.[inout] vhtot: The integrated mixed layer meridional velocity, H m s-1.[inout] r0_tot: The integrated mixed layer potential density referenced to 0 pressure [H kg m-3 ~> kg m-2 or kg2 m-5].[inout] rcv_tot: The integrated mixed layer coordinate variable potential density [H kg m-3 ~> kg m-2 or kg2 m-5].[in] u: Zonal velocities interpolated to h points, m s-1.[in] v: Zonal velocities interpolated to h points, m s-1.[in] t: Layer temperatures [degC].[in] s: Layer salinities [ppt].[in] r0: Potential density referenced to[in] rcv: The coordinate defining potential[in] eps: The negligibly small amount of water[in] dr0_dt: The partial derivative of R0 with respect to temperature [kg m-3 degC-1].[in] drcv_dt: The partial derivative of Rcv with respect to temperature [kg m-3 degC-1].[in] cmke: Coefficients of HpE and HpE^2 used in calculating the denominator of MKE_rate; the two elements have differing units of [H-1 ~> m-1 or m2 kg-1] and [H-2 ~> m-2 or m4 kg-2].[in] idt_diag: The inverse of the accumulated diagnostic time interval [T-1 ~> s-1].[in] nsw: The number of bands of penetrating shortwave radiation.[inout] pen_sw_bnd: The penetrating shortwave heating at the sea surface in each penetrating band [degC H ~> degC m or degC kg m-2].[in] opacity_band: The opacity in each band of penetrating shortwave radiation [H-1 ~> m-1 or m2 kg-1].[inout] tke: The turbulent kinetic energy available for mixing over a time step [Z m2 T-2 ~> m3 s-2].[inout] idecay_len_tke: The vertical TKE decay rate [H-1 ~> m-1 or m2 kg-1].[in] j: The j-index to work on.[in] ksort: The density-sorted k-indicies.cs: The control structure for this module.
-
subroutine
sort_ml(h h, R0 R0, eps eps, G G, GV GV, CS CS, ksort ksort)¶ This subroutine generates an array of indices that are sorted by layer density.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] h: Layer thickness [H ~> m or kg m-2].[in] r0: The potential density used to sort the layers [kg m-3].[in] eps: The (small) thickness that must remain in each layer [H ~> m or kg m-2].cs: The control structure returned by a previous call to mixedlayer_init.[out] ksort: The k-index to use in the sort.
-
subroutine
resort_ml(h h, T T, S S, R0 R0, Rcv Rcv, RcvTgt RcvTgt, eps eps, d_ea d_ea, d_eb d_eb, ksort ksort, G G, GV GV, CS CS, dR0_dT dR0_dT, dR0_dS dR0_dS, dRcv_dT dRcv_dT, dRcv_dS dRcv_dS)¶ This subroutine actually moves properties between layers to achieve a resorted state, with all of the resorted water either moved into the correct interior layers or in the top nkmb layers.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[inout] h: Layer thickness [H ~> m or kg m-2]. Layer 0 is the new mixed layer.[inout] t: Layer temperatures [degC].[inout] s: Layer salinities [ppt].[inout] r0: Potential density referenced to surface pressure [kg m-3].[inout] rcv: The coordinate defining potential density [kg m-3].[in] rcvtgt: The target value of Rcv for each layer [kg m-3].[inout] eps: The (small) thickness that must remain in each layer [H ~> m or kg m-2].[inout] d_ea: The upward increase across a layer in the entrainment from above [H ~> m or kg m-2]. Positive d_ea goes with layer thickness increases.[inout] d_eb: The downward increase across a layer in the entrainment from below [H ~> m or kg m-2]. Positive values go with mass gain by a layer.[in] ksort: The density-sorted k-indicies.cs: The control structure for this module.[in] dr0_dt: The partial derivative of potential density referenced to the surface with potential temperature [kg m-3 degC-1].[in] dr0_ds: The partial derivative of cpotential density referenced to the surface with salinity, [kg m-3 ppt-1].[in] drcv_dt: The partial derivative of coordinate defining potential density with potential temperature [kg m-3 degC-1].[in] drcv_ds: The partial derivative of coordinate defining potential density with salinity, [kg m-3 ppt-1].
-
subroutine
mixedlayer_detrain_2(h h, T T, S S, R0 R0, Rcv Rcv, RcvTgt RcvTgt, dt_in_T dt_in_T, dt_diag dt_diag, d_ea d_ea, j j, G G, GV GV, US US, CS CS, dR0_dT dR0_dT, dR0_dS dR0_dS, dRcv_dT dRcv_dT, dRcv_dS dRcv_dS, max_BL_det max_BL_det)¶ This subroutine moves any water left in the former mixed layers into the two buffer layers and may also move buffer layer water into the interior isopycnal layers.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[inout] h: Layer thickness [H ~> m or kg m-2]. Layer 0 is the new mixed layer.[inout] t: Potential temperature [degC].[inout] s: Salinity [ppt].[inout] r0: Potential density referenced to surface pressure [kg m-3].[inout] rcv: The coordinate defining potential density [kg m-3].[in] rcvtgt: The target value of Rcv for each layer [kg m-3].[in] dt_in_t: Time increment [T ~> s].[in] dt_diag: The diagnostic time step [T ~> s].[inout] d_ea: The upward increase across a layer in the entrainment from above [H ~> m or kg m-2]. Positive d_ea goes with layer thickness increases.[in] j: The meridional row to work on.[in] us: A dimensional unit scaling typecs: The control structure returned by a previous call to mixedlayer_init.[in] dr0_dt: The partial derivative of potential density referenced to the surface with potential temperature, [kg m-3 degC-1].[in] dr0_ds: The partial derivative of cpotential density referenced to the surface with salinity [kg m-3 ppt-1].[in] drcv_dt: The partial derivative of coordinate defining potential density with potential temperature, [kg m-3 degC-1].[in] drcv_ds: The partial derivative of coordinate defining potential density with salinity [kg m-3 ppt-1].[in] max_bl_det: If non-negative, the maximum detrainment permitted from the buffer layers [H ~> m or kg m-2].
-
subroutine
mixedlayer_detrain_1(h h, T T, S S, R0 R0, Rcv Rcv, RcvTgt RcvTgt, dt_in_T dt_in_T, dt_diag dt_diag, d_ea d_ea, d_eb d_eb, j j, G G, GV GV, US US, CS CS, dRcv_dT dRcv_dT, dRcv_dS dRcv_dS, max_BL_det max_BL_det)¶ This subroutine moves any water left in the former mixed layers into the single buffer layers and may also move buffer layer water into the interior isopycnal layers.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[inout] h: Layer thickness [H ~> m or kg m-2]. Layer 0 is the new mixed layer.[inout] t: Potential temperature [degC].[inout] s: Salinity [ppt].[inout] r0: Potential density referenced to surface pressure [kg m-3].[inout] rcv: The coordinate defining potential density [kg m-3].[in] rcvtgt: The target value of Rcv for each layer [kg m-3].[in] dt_in_t: Time increment [T ~> s].[in] dt_diag: The accumulated time interval for diagnostics [T ~> s].[inout] d_ea: The upward increase across a layer in the entrainment from above [H ~> m or kg m-2]. Positive d_ea goes with layer thickness increases.[inout] d_eb: The downward increase across a layer in the entrainment from below [H ~> m or kg m-2]. Positive values go with mass gain by a layer.[in] j: The meridional row to work on.[in] us: A dimensional unit scaling typecs: The control structure returned by a previous call to mixedlayer_init.[in] drcv_dt: The partial derivative of coordinate defining potential density with potential temperature [kg m-3 degC-1].[in] drcv_ds: The partial derivative of coordinate defining potential density with salinity [kg m-3 ppt-1].[in] max_bl_det: If non-negative, the maximum detrainment permitted from the buffer layers [H ~> m or kg m-2].
-
subroutine, public mom_bulk_mixed_layer::bulkmixedlayer_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS) This subroutine initializes the MOM bulk mixed layer module.
- Parameters
[in] time: The model’s clock with the current time.[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters.[inout] diag: A structure that is used to regulate diagnostic output.cs: A pointer that is set to point to the control structure for this module.
-
real function mom_bulk_mixed_layer::ef4(Ht Ht, En En, I_L I_L, dR_de dR_de) This subroutine returns an approximation to the integral R = exp(-L*(H+E)) integral(LH to L(H+E)) L/(1-(1+x)exp(-x)) dx. The approximation to the integrand is good to within -2% at x~.3 and +25% at x~3.5, but the exponential deemphasizes the importance of large x. When L=0, EF4 returns E/((Ht+E)*Ht).
- Return
The integral [H-1 ~> m-1 or m2 kg-1].
- Parameters
[in] ht: Total thickness [H ~> m or kg m-2].[in] en: Entrainment [H ~> m or kg m-2].[in] i_l: The e-folding scale [H-1 ~> m-1 or m2 kg-1][inout] dr_de: The partial derivative of the result R with E [H-2 ~> m-2 or m4 kg-2].
-
integer
-
namespace
mom_checksum_packages¶ Provides routines that do checksums of groups of MOM variables.
Functions
-
subroutine
mom_state_chksum_5arg(mesg mesg, u u, v v, h h, uh uh, vh vh, G G, GV GV, haloshift haloshift, symmetric symmetric)¶ Write out chksums for the model’s basic state variables, including transports.
- Parameters
[in] mesg: A message that appears on the chksum lines.[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] u: The zonal velocity [m s-1].[in] v: The meridional velocity [m s-1].[in] h: Layer thicknesses [H ~> m or kg m-2].[in] uh: Volume flux through zonal faces = u*h*dy[in] vh: Volume flux through meridional faces = v*h*dx[in] haloshift: The width of halos to check (default 0).[in] symmetric: If true, do checksums on the fully symmetric computationoal domain.
-
subroutine
mom_state_chksum_3arg(mesg mesg, u u, v v, h h, G G, GV GV, haloshift haloshift, symmetric symmetric)¶ Write out chksums for the model’s basic state variables.
- Parameters
[in] mesg: A message that appears on the chksum lines.[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] u: Zonal velocity [m s-1].[in] v: Meridional velocity [m s-1].[in] h: Layer thicknesses [H ~> m or kg m-2].[in] haloshift: The width of halos to check (default 0).[in] symmetric: If true, do checksums on the fully symmetric computationoal domain.
-
subroutine, public mom_checksum_packages::mom_thermo_chksum(mesg mesg, tv tv, G G, haloshift haloshift) Write out chksums for the model’s thermodynamic state variables.
- Parameters
[in] mesg: A message that appears on the chksum lines.[in] tv: A structure pointing to various thermodynamic variables.[in] g: The ocean’s grid structure.[in] haloshift: The width of halos to check (default 0).
-
subroutine, public mom_checksum_packages::mom_surface_chksum(mesg mesg, sfc sfc, G G, haloshift haloshift, symmetric symmetric) Write out chksums for the ocean surface variables.
- Parameters
[in] mesg: A message that appears on the chksum lines.[inout] sfc: transparent ocean surface state structure shared with the calling routine data in this structure is intent out.[in] g: The ocean’s grid structure.[in] haloshift: The width of halos to check (default 0).[in] symmetric: If true, do checksums on the fully symmetric computationoal domain.
-
subroutine, public mom_checksum_packages::mom_accel_chksum(mesg mesg, CAu CAu, CAv CAv, PFu PFu, PFv PFv, diffu diffu, diffv diffv, G G, GV GV, US US, pbce pbce, u_accel_bt u_accel_bt, v_accel_bt v_accel_bt, symmetric symmetric) Write out chksums for the model’s accelerations.
- Parameters
[in] mesg: A message that appears on the chksum lines.[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] cau: Zonal acceleration due to Coriolis[in] cav: Meridional acceleration due to Coriolis[in] pfu: Zonal acceleration due to pressure gradients[in] pfv: Meridional acceleration due to pressure gradients[in] diffu: Zonal acceleration due to convergence of the[in] diffv: Meridional acceleration due to convergence of[in] us: A dimensional unit scaling type[in] pbce: The baroclinic pressure anomaly in each layer[in] u_accel_bt: The zonal acceleration from terms in the[in] v_accel_bt: The meridional acceleration from terms in[in] symmetric: If true, do checksums on the fully symmetric computationoal domain.
-
subroutine, public mom_checksum_packages::mom_state_stats(mesg mesg, u u, v v, h h, Temp Temp, Salt Salt, G G, allowChange allowChange, permitDiminishing permitDiminishing) Monitor and write out statistics for the model’s state variables.
- Parameters
[in] g: The ocean’s grid structure.[in] mesg: A message that appears on the chksum lines.[in] u: The zonal velocity [m s-1].[in] v: The meridional velocity [m s-1].[in] h: Layer thicknesses [H ~> m or kg m-2].[in] temp: Temperature [degC].[in] salt: Salinity [ppt].[in] allowchange: do not flag an error if the statistics change.[in] permitdiminishing: do not flag error if the extrema are diminishing.
-
subroutine
-
namespace
mom_checksums¶ Routines to calculate checksums of various array and vector types.
Functions
-
subroutine, public mom_checksums::chksum0(scalar scalar, mesg mesg, scale scale, logunit logunit) Checksum a scalar field (consistent with array checksums)
- Parameters
[in] scalar: The array to be checksummed[in] mesg: An identifying message[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine, public mom_checksums::zchksum(array array, mesg mesg, scale scale, logunit logunit) Checksum a 1d array (typically a column).
- Parameters
[in] array: The array to be checksummed[in] mesg: An identifying message[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
chksum_pair_h_2d(mesg mesg, arrayA arrayA, arrayB arrayB, HI HI, haloshift haloshift, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums on a pair of 2d arrays staggered at tracer points.
- Parameters
[in] mesg: Identifying messages[in] hi: A horizontal index type[in] arraya: The first array to be checksummed[in] arrayb: The second array to be checksummed[in] haloshift: The width of halos to check (default 0)[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
chksum_pair_h_3d(mesg mesg, arrayA arrayA, arrayB arrayB, HI HI, haloshift haloshift, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums on a pair of 3d arrays staggered at tracer points.
- Parameters
[in] mesg: Identifying messages[in] hi: A horizontal index type[in] arraya: The first array to be checksummed[in] arrayb: The second array to be checksummed[in] haloshift: The width of halos to check (default 0)[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
chksum_h_2d(array array, mesg mesg, HI HI, haloshift haloshift, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums a 2d array staggered at tracer points.
- Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] mesg: An identifying message[in] haloshift: The width of halos to check (default 0)[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
chksum_pair_b_2d(mesg mesg, arrayA arrayA, arrayB arrayB, HI HI, haloshift haloshift, symmetric symmetric, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums on a pair of 2d arrays staggered at q-points.
- Parameters
[in] mesg: Identifying messages[in] hi: A horizontal index type[in] arraya: The first array to be checksummed[in] arrayb: The second array to be checksummed[in] symmetric: If true, do the checksums on the full symmetric computational domain.[in] haloshift: The width of halos to check (default 0)[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
chksum_pair_b_3d(mesg mesg, arrayA arrayA, arrayB arrayB, HI HI, haloshift haloshift, symmetric symmetric, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums on a pair of 3d arrays staggered at q-points.
- Parameters
[in] mesg: Identifying messages[in] hi: A horizontal index type[in] arraya: The first array to be checksummed[in] arrayb: The second array to be checksummed[in] haloshift: The width of halos to check (default 0)[in] symmetric: If true, do the checksums on the full symmetric computational domain.[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
chksum_b_2d(array array, mesg mesg, HI HI, haloshift haloshift, symmetric symmetric, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums a 2d array staggered at corner points.
- Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] mesg: An identifying message[in] haloshift: The width of halos to check (default 0)[in] symmetric: If true, do the checksums on the full symmetric computational domain.[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
chksum_uv_2d(mesg mesg, arrayU arrayU, arrayV arrayV, HI HI, haloshift haloshift, symmetric symmetric, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums a pair of 2d velocity arrays staggered at C-grid locations.
- Parameters
[in] mesg: Identifying messages[in] hi: A horizontal index type[in] arrayu: The u-component array to be checksummed[in] arrayv: The v-component array to be checksummed[in] haloshift: The width of halos to check (default 0)[in] symmetric: If true, do the checksums on the full symmetric computational domain.[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for these arrays.[in] logunit: IO unit for checksum logging
-
subroutine
chksum_uv_3d(mesg mesg, arrayU arrayU, arrayV arrayV, HI HI, haloshift haloshift, symmetric symmetric, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums a pair of 3d velocity arrays staggered at C-grid locations.
- Parameters
[in] mesg: Identifying messages[in] hi: A horizontal index type[in] arrayu: The u-component array to be checksummed[in] arrayv: The v-component array to be checksummed[in] haloshift: The width of halos to check (default 0)[in] symmetric: If true, do the checksums on the full symmetric computational domain.[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for these arrays.[in] logunit: IO unit for checksum logging
-
subroutine
chksum_u_2d(array array, mesg mesg, HI HI, haloshift haloshift, symmetric symmetric, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums a 2d array staggered at C-grid u points.
- Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] mesg: An identifying message[in] haloshift: The width of halos to check (default 0)[in] symmetric: If true, do the checksums on the full symmetric computational domain.[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
chksum_v_2d(array array, mesg mesg, HI HI, haloshift haloshift, symmetric symmetric, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums a 2d array staggered at C-grid v points.
- Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] mesg: An identifying message[in] haloshift: The width of halos to check (default 0)[in] symmetric: If true, do the checksums on the full symmetric computational domain.[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
chksum_h_3d(array array, mesg mesg, HI HI, haloshift haloshift, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums a 3d array staggered at tracer points.
- Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] mesg: An identifying message[in] haloshift: The width of halos to check (default 0)[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
chksum_b_3d(array array, mesg mesg, HI HI, haloshift haloshift, symmetric symmetric, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums a 3d array staggered at corner points.
- Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] mesg: An identifying message[in] haloshift: The width of halos to check (default 0)[in] symmetric: If true, do the checksums on the full symmetric computational domain.[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
chksum_u_3d(array array, mesg mesg, HI HI, haloshift haloshift, symmetric symmetric, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums a 3d array staggered at C-grid u points.
- Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] mesg: An identifying message[in] haloshift: The width of halos to check (default 0)[in] symmetric: If true, do the checksums on the full symmetric computational domain.[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
chksum_v_3d(array array, mesg mesg, HI HI, haloshift haloshift, symmetric symmetric, omit_corners omit_corners, scale scale, logunit logunit)¶ Checksums a 3d array staggered at C-grid v points.
- Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] mesg: An identifying message[in] haloshift: The width of halos to check (default 0)[in] symmetric: If true, do the checksums on the full symmetric computational domain.[in] omit_corners: If true, avoid checking diagonal shifts[in] scale: A scaling factor for this array.[in] logunit: IO unit for checksum logging
-
subroutine
chksum1d(array array, mesg mesg, start_i start_i, end_i end_i, compare_PEs compare_PEs)¶ chksum1d does a checksum of a 1-dimensional array.
- Parameters
[in] array: The array to be summed (index starts at 1).[in] mesg: An identifying message.[in] start_i: The starting index for the sum (default 1)[in] end_i: The ending index for the sum (default all)[in] compare_pes: If true, compare across PEs instead of summing and list the root_PE value (default true)
-
subroutine
chksum2d(array array, mesg mesg)¶ chksum2d does a checksum of all data in a 2-d array.
- Parameters
array: The array to be checksummedmesg: An identifying message
-
subroutine
chksum3d(array array, mesg mesg)¶ chksum3d does a checksum of all data in a 2-d array.
- Parameters
array: The array to be checksummedmesg: An identifying message
-
logical function mom_checksums::is_nan_0d(x x) This function returns .true. if x is a NaN, and .false. otherwise.
- Parameters
[in] x: The value to be checked for NaNs.
-
logical function mom_checksums::is_nan_1d(x x, skip_mpp skip_mpp) Returns .true. if any element of x is a NaN, and .false. otherwise.
- Parameters
[in] x: The array to be checked for NaNs.[in] skip_mpp: If true, only check this array only on the local PE (default false).
-
logical function mom_checksums::is_nan_2d(x x) Returns .true. if any element of x is a NaN, and .false. otherwise.
- Parameters
[in] x: The array to be checked for NaNs.
-
logical function mom_checksums::is_nan_3d(x x) Returns .true. if any element of x is a NaN, and .false. otherwise.
- Parameters
[in] x: The array to be checked for NaNs.
-
subroutine
chk_sum_msg1(fmsg fmsg, bc0 bc0, mesg mesg, iounit iounit)¶ Write a message including the checksum of the non-shifted array.
- Parameters
[in] fmsg: A checksum code-location specific preamble[in] mesg: An identifying message supplied by top-level caller[in] bc0: The bitcount of the non-shifted array[in] iounit: Checksum logger IO unit
-
subroutine
chk_sum_msg5(fmsg fmsg, bc0 bc0, bcSW bcSW, bcSE bcSE, bcNW bcNW, bcNE bcNE, mesg mesg, iounit iounit)¶ Write a message including checksums of non-shifted and diagonally shifted arrays.
- Parameters
[in] fmsg: A checksum code-location specific preamble[in] mesg: An identifying message supplied by top-level caller[in] bc0: The bitcount of the non-shifted array[in] bcsw: The bitcount for SW shifted array[in] bcse: The bitcount for SE shifted array[in] bcnw: The bitcount for NW shifted array[in] bcne: The bitcount for NE shifted array[in] iounit: Checksum logger IO unit
-
subroutine
chk_sum_msg_nsew(fmsg fmsg, bc0 bc0, bcN bcN, bcS bcS, bcE bcE, bcW bcW, mesg mesg, iounit iounit)¶ Write a message including checksums of non-shifted and laterally shifted arrays.
- Parameters
[in] fmsg: A checksum code-location specific preamble[in] mesg: An identifying message supplied by top-level caller[in] bc0: The bitcount of the non-shifted array[in] bcn: The bitcount for N shifted array[in] bcs: The bitcount for S shifted array[in] bce: The bitcount for E shifted array[in] bcw: The bitcount for W shifted array[in] iounit: Checksum logger IO unit
-
subroutine
chk_sum_msg_s(fmsg fmsg, bc0 bc0, bcS bcS, mesg mesg, iounit iounit)¶ Write a message including checksums of non-shifted and southward shifted arrays.
- Parameters
[in] fmsg: A checksum code-location specific preamble[in] mesg: An identifying message supplied by top-level caller[in] bc0: The bitcount of the non-shifted array[in] bcs: The bitcount of the south-shifted array[in] iounit: Checksum logger IO unit
-
subroutine
chk_sum_msg_w(fmsg fmsg, bc0 bc0, bcW bcW, mesg mesg, iounit iounit)¶ Write a message including checksums of non-shifted and westward shifted arrays.
- Parameters
[in] fmsg: A checksum code-location specific preamble[in] mesg: An identifying message supplied by top-level caller[in] bc0: The bitcount of the non-shifted array[in] bcw: The bitcount of the west-shifted array[in] iounit: Checksum logger IO unit
-
subroutine
chk_sum_msg2(fmsg fmsg, bc0 bc0, bcSW bcSW, mesg mesg, iounit iounit)¶ Write a message including checksums of non-shifted and southwestward shifted arrays.
- Parameters
[in] fmsg: A checksum code-location specific preamble[in] mesg: An identifying message supplied by top-level caller[in] bc0: The bitcount of the non-shifted array[in] bcsw: The bitcount of the southwest-shifted array[in] iounit: Checksum logger IO unit
-
subroutine
chk_sum_msg3(fmsg fmsg, aMean aMean, aMin aMin, aMax aMax, mesg mesg, iounit iounit)¶ Write a message including the global mean, maximum and minimum of an array.
- Parameters
[in] fmsg: A checksum code-location specific preamble[in] mesg: An identifying message supplied by top-level caller[in] amean: The mean value of the array[in] amin: The minimum value of the array[in] amax: The maximum value of the array[in] iounit: Checksum logger IO unit
-
subroutine, public mom_checksums::mom_checksums_init(param_file param_file) MOM_checksums_init initializes the MOM_checksums module. As it happens, the only thing that it does is to log the version of this module.
- Parameters
[in] param_file: A structure to parse for run-time parameters
-
subroutine
chksum_error(signal signal, message message)¶ A wrapper for MOM_error used in the checksum code.
- Parameters
[in] signal: An error severity level, such as FATAL or WARNING[in] message: An error message
-
integer function mom_checksums::bitcount(x x) Does a bitcount of a number by first casting to an integer and then using BTEST to check bit by bit.
- Parameters
x: Number to be bitcount
Variables
-
integer, parameter mom_checksums::bc_modulus = 1000000000 Modulus of checksum bitcount.
-
integer, parameter mom_checksums::default_shift =0 The default array shift.
-
logical
calculatestatistics= .true.¶ If true, report min, max and mean.
-
logical
writechksums= .true.¶ If true, report the bitcount checksum.
-
logical
checkfornans= .true.¶ If true, checks array for NaNs and cause FATAL error is any are found.
-
-
namespace
mom_coms¶ Interfaces to non-domain-oriented communication subroutines, including the MOM6 reproducing sums facility.
Functions
-
real function mom_coms::reproducing_sum_2d(array array, isr isr, ier ier, jsr jsr, jer jer, EFP_sum EFP_sum, reproducing reproducing, overflow_check overflow_check, err err) This subroutine uses a conversion to an integer representation of real numbers to give an order-invariant sum of distributed 2-D arrays that reproduces across domain decomposition. This technique is described in Hallberg & Adcroft, 2014, Parallel Computing, doi:10.1016/j.parco.2014.04.007.
- Return
Result
- Parameters
[in] array: The array to be summed[in] isr: The starting i-index of the sum, noting that the array indices starts at 1[in] ier: The ending i-index of the sum, noting that the array indices starts at 1[in] jsr: The starting j-index of the sum, noting that the array indices starts at 1[in] jer: The ending j-index of the sum, noting that the array indices starts at 1[out] efp_sum: The result in extended fixed point format[in] reproducing: If present and false, do the sum using the naive non-reproducing approach[in] overflow_check: If present and false, disable checking for overflows in incremental results. This can speed up calculations if the number of values being summed is small enough[out] err: If present, return an error code instead of triggering any fatal errors directly from this routine.
-
real function mom_coms::reproducing_sum_3d(array array, isr isr, ier ier, jsr jsr, jer jer, sums sums, EFP_sum EFP_sum, err err) This subroutine uses a conversion to an integer representation of real numbers to give an order-invariant sum of distributed 3-D arrays that reproduces across domain decomposition. This technique is described in Hallberg & Adcroft, 2014, Parallel Computing, doi:10.1016/j.parco.2014.04.007.
- Return
Result
- Parameters
[in] array: The array to be summed[in] isr: The starting i-index of the sum, noting that the array indices starts at 1[in] ier: The ending i-index of the sum, noting that the array indices starts at 1[in] jsr: The starting j-index of the sum, noting that the array indices starts at 1[in] jer: The ending j-index of the sum, noting that the array indices starts at 1[out] sums: The sums by vertical layer[out] efp_sum: The result in extended fixed point format[out] err: If present, return an error code instead of triggering any fatal errors directly from this routine.
-
integer(kind=8) function, dimension(ni) mom_coms::real_to_ints(r r, prec_error prec_error, overflow overflow) Convert a real number into the array of integers constitute its extended-fixed-point representation.
- Parameters
[in] r: The real number being converted[in] prec_error: The PE-count dependent precision of the integers that is safe from overflows during global sums. This will be larger than the compile-time precision parameter, and is used to detect overflows.[inout] overflow: Returns true if the conversion is being done on a value that is too large to be represented
-
real function mom_coms::ints_to_real(ints ints) Convert the array of integers that constitute an extended-fixed-point representation into a real number.
- Parameters
[in] ints: The array of EFP integers
-
subroutine
increment_ints(int_sum int_sum, int2 int2, prec_error prec_error)¶ Increment an array of integers that constitutes an extended-fixed-point representation with a another EFP number.
- Parameters
[inout] int_sum: The array of EFP integers being incremented[in] int2: The array of EFP integers being added[in] prec_error: The PE-count dependent precision of the integers that is safe from overflows during global sums. This will be larger than the compile-time precision parameter, and is used to detect overflows.
-
subroutine
increment_ints_faster(int_sum int_sum, r r, max_mag_term max_mag_term)¶ Increment an EFP number with a real number without doing any carrying of of overflows and using only minimal error checking.
- Parameters
[inout] int_sum: The array of EFP integers being incremented[in] r: The real number being added.[inout] max_mag_term: A running maximum magnitude of the r’s.
-
subroutine
carry_overflow(int_sum int_sum, prec_error prec_error)¶ This subroutine handles carrying of the overflow.
- Parameters
[inout] int_sum: The array of EFP integers being modified by carries, but without changing value.[in] prec_error: The PE-count dependent precision of the integers that is safe from overflows during global sums. This will be larger than the compile-time precision parameter, and is used to detect overflows.
-
subroutine
regularize_ints(int_sum int_sum)¶ This subroutine carries the overflow, and then makes sure that all integers are of the same sign as the overall value.
- Parameters
[inout] int_sum: The array of integers being modified to take a
-
logical function, public mom_coms::query_efp_overflow_error() Returns the status of the module’s error flag.
-
subroutine, public mom_coms::reset_efp_overflow_error() Reset the module’s error flag to false.
-
type(efp_type) function, public mom_coms::efp_plus(EFP1 EFP1, EFP2 EFP2) Add two extended-fixed-point numbers.
- Return
The result in extended fixed point format
- Parameters
[in] efp1: The first extended fixed point number[in] efp2: The second extended fixed point number
-
type(efp_type) function, public mom_coms::efp_minus(EFP1 EFP1, EFP2 EFP2) Subract one extended-fixed-point number from another.
- Return
The result in extended fixed point format
- Parameters
[in] efp1: The first extended fixed point number[in] efp2: The extended fixed point number being subtracted from the first extended fixed point number
-
subroutine
efp_assign(EFP1 EFP1, EFP2 EFP2)¶ Copy one extended-fixed-point number into another.
- Parameters
[out] efp1: The recipient extended fixed point number[in] efp2: The source extended fixed point number
-
real function, public mom_coms::efp_to_real(EFP1 EFP1) Return the real number that an extended-fixed-point number corresponds with.
- Parameters
[inout] efp1: The extended fixed point number being converted
-
real function, public mom_coms::efp_real_diff(EFP1 EFP1, EFP2 EFP2) Take the difference between two extended-fixed-point numbers (EFP1 - EFP2) and return the result as a real number.
- Return
The real result
- Parameters
[in] efp1: The first extended fixed point number[in] efp2: The extended fixed point number being subtracted from the first extended fixed point number
-
type(efp_type) function, public mom_coms::real_to_efp(val val, overflow overflow) Return the extended-fixed-point number that a real number corresponds with.
- Parameters
[in] val: The real number being converted[inout] overflow: Returns true if the conversion is being done on a value that is too large to be represented
-
subroutine, public mom_coms::efp_list_sum_across_pes(EFPs EFPs, nval nval, errors errors) This subroutine does a sum across PEs of a list of EFP variables, returning the sums in place, with all overflows carried.
- Parameters
[inout] efps: The list of extended fixed point numbers[in] nval: The number of values being summed.[out] errors: A list of error flags for each sum
-
subroutine, public mom_coms::mom_infra_end() This subroutine carries out all of the calls required to close out the infrastructure cleanly. This should only be called in ocean-only runs, as the coupler takes care of this in coupled runs.
Variables
-
integer(kind=8), parameter mom_coms::prec =2_8**46 The precision of each integer.
-
real, parameter mom_coms::r_prec =2.0**46 A real version of prec.
-
real, parameter mom_coms::i_prec =1.0/(2.0**46) The inverse of prec.
-
integer, parameter mom_coms::max_count_prec =2**(63-46)-1 The number of values that can be added together with the current value of prec before there will be roundoff problems.
-
integer, parameter mom_coms::ni =6 The number of long integers to use to represent a real number.
-
real, dimension(ni), parameter mom_coms::pr = (/ r_prec**2, r_prec, 1.0, 1.0/r_prec, 1.0/r_prec**2, 1.0/r_prec**3 /) An array of the real precision of each of the integers.
-
real, dimension(ni), parameter mom_coms::i_pr = (/ 1.0/r_prec**2, 1.0/r_prec, 1.0, r_prec, r_prec**2, r_prec**3 /) An array of the inverse of the real precision of each of the integers.
-
real, parameter mom_coms::max_efp_float = pr(1) * (2.**63 - 1.) The largest float with an EFP representation. NOTE: Only the first bin can exceed precision, but is bounded by the largest signed integer.
-
logical
overflow_error= .false.¶ This becomes true if an overflow is encountered.
-
logical
nan_error= .false.¶ This becomes true if a NaN is encountered.
-
logical
debug= .false.¶ Making this true enables debugging output.
-
-
namespace
mom_constants¶ Provides a few physical constants.
Variables
-
real, parameter, public mom_constants::celsius_kelvin_offset = 273.15 The constant offset for converting temperatures in Kelvin to Celsius.
-
-
namespace
mom_continuity¶ Solve the layer continuity equation.
Functions
-
subroutine, public mom_continuity::continuity(u u, v v, hin hin, h h, uh uh, vh vh, dt dt, G G, GV GV, US US, CS CS, uhbt uhbt, vhbt vhbt, OBC OBC, visc_rem_u visc_rem_u, visc_rem_v visc_rem_v, u_cor u_cor, v_cor v_cor, uhbt_aux uhbt_aux, vhbt_aux vhbt_aux, u_cor_aux u_cor_aux, v_cor_aux v_cor_aux, BT_cont BT_cont) Time steps the layer thicknesses, using a monotonically limited, directionally split PPM scheme, based on Lin (1994).
- Parameters
[inout] g: Ocean grid structure.[in] gv: Vertical grid structure.[in] u: Zonal velocity [m s-1].[in] v: Meridional velocity [m s-1].[in] hin: Initial layer thickness [H ~> m or kg m-2].[inout] h: Final layer thickness [H ~> m or kg m-2].[out] uh: Volume flux through zonal faces =[out] vh: Volume flux through meridional faces =[in] dt: Time increment [s].[in] us: A dimensional unit scaling typecs: Control structure for mom_continuity.[in] uhbt: The vertically summed volume[in] vhbt: The vertically summed volumeobc: Open boundaries control structure.[in] visc_rem_u: Both the fraction of[in] visc_rem_v: Both the fraction of[out] u_cor: The zonal velocities that[out] v_cor: The meridional velocities that[in] uhbt_aux: A second summed zonal[in] vhbt_aux: A second summed meridional[inout] u_cor_aux: The zonal velocities[inout] v_cor_aux: The meridional velocitiesbt_cont: A structure with elements
-
subroutine, public mom_continuity::continuity_init(Time Time, G G, GV GV, param_file param_file, diag diag, CS CS) Initializes continuity_cs.
- Parameters
[in] time: Current model time.[in] g: Ocean grid structure.[in] gv: Vertical grid structure.[in] param_file: Parameter file handles.[inout] diag: Diagnostics control structure.cs: Control structure for mom_continuity.
-
integer function, public mom_continuity::continuity_stencil(CS CS) continuity_stencil returns the continuity solver stencil size
- Return
The continuity solver stencil size with the current settings.
- Parameters
cs: Module’s control structure.
-
subroutine, public mom_continuity::continuity_end(CS CS) Destructor for continuity_cs.
- Parameters
cs: Control structure for mom_continuity.
Variables
-
integer, parameter mom_continuity::ppm_scheme = 1 Enumerated constant to select PPM.
-
character(len=20), parameter mom_continuity::ppm_string = "PPM" String to select PPM.
-
-
namespace
mom_continuity_ppm¶ Solve the layer continuity equation using the PPM method for layer fluxes.
This module contains the subroutines that advect layer thickness. The scheme here uses a Piecewise-Parabolic method with a positive definite limiter.
Unnamed Group
-
integer
id_clock_update¶ CPU time clock IDs.
-
integer
id_clock_correct¶ CPU time clock IDs.
-
subroutine, public mom_continuity_ppm::continuity_ppm(u u, v v, hin hin, h h, uh uh, vh vh, dt dt, G G, GV GV, US US, CS CS, uhbt uhbt, vhbt vhbt, OBC OBC, visc_rem_u visc_rem_u, visc_rem_v visc_rem_v, u_cor u_cor, v_cor v_cor, uhbt_aux uhbt_aux, vhbt_aux vhbt_aux, u_cor_aux u_cor_aux, v_cor_aux v_cor_aux, BT_cont BT_cont) Time steps the layer thicknesses, using a monotonically limit, directionally split PPM scheme, based on Lin (1994).
- Parameters
[inout] g: The ocean’s grid structure.cs: Module’s control structure.[in] u: Zonal velocity [m s-1].[in] v: Meridional velocity [m s-1].[in] hin: Initial layer thickness [H ~> m or kg m-2].[inout] h: Final layer thickness [H ~> m or kg m-2].[out] uh: Zonal volume flux, u*h*dy [H m2 s-1 ~> m3 s-1 or kg s-1].[out] vh: Meridional volume flux, v*h*dx [H m2 s-1 ~> m3 s-1 or kg s-1].[in] dt: Time increment [s].[in] gv: Vertical grid structure.[in] us: A dimensional unit scaling type[in] uhbt: The summed volume flux through zonal faces[in] vhbt: The summed volume flux through meridional facesobc: Open boundaries control structure.[in] visc_rem_u: The fraction of zonal momentum originally in a layer that remains after a time-step of viscosity, and the fraction of a time-step’s worth of a barotropic acceleration that a layer experiences after viscosity is applied. Non-dimensional between 0 (at the bottom) and 1 (far above the bottom).[in] visc_rem_v: The fraction of meridional momentum originally in a layer that remains after a time-step of viscosity, and the fraction of a time-step’s worth of a barotropic acceleration that a layer experiences after viscosity is applied. Non-dimensional between 0 (at the bottom) and 1 (far above the bottom).[out] u_cor: The zonal velocities that give uhbt as the depth-integrated transport [m s-1].[out] v_cor: The meridional velocities that give vhbt as the depth-integrated transport [m s-1].[in] uhbt_aux: A second set of summed volume fluxes through zonal faces [H m2 s-1 ~> m3 s-1 or kg s-1].[in] vhbt_aux: A second set of summed volume fluxes through meridional faces [H m2 s-1 ~> m3 s-1 or kg s-1].[out] u_cor_aux: The zonal velocities that give uhbt_aux as the depth-integrated transports [m s-1].[out] v_cor_aux: The meridional velocities that give vhbt_aux as the depth-integrated transports [m s-1].bt_cont: A structure with elements that describe the effective open face areas as a function of barotropic flow.
-
subroutine
zonal_mass_flux(u u, h_in h_in, uh uh, dt dt, G G, GV GV, US US, CS CS, LB LB, uhbt uhbt, OBC OBC, visc_rem_u visc_rem_u, u_cor u_cor, uhbt_aux uhbt_aux, u_cor_aux u_cor_aux, BT_cont BT_cont)¶ Calculates the mass or volume fluxes through the zonal faces, and other related quantities.
- Parameters
[inout] g: Ocean’s grid structure.[in] gv: Ocean’s vertical grid structure.[in] u: Zonal velocity [m s-1].[in] h_in: Layer thickness used to calculate fluxes [H ~> m or kg m-2].[out] uh: Volume flux through zonal faces = u*h*dy[in] dt: Time increment [s].[in] us: A dimensional unit scaling typecs: This module’s control structure.[in] lb: Loop bounds structure.obc: Open boundaries control structure.[in] visc_rem_u: The fraction of zonal momentum originally in a layer that remains after a time-step of viscosity, and the fraction of a time-step’s worth of a barotropic acceleration that a layer experiences after viscosity is applied. Non-dimensional between 0 (at the bottom) and 1 (far above the bottom).[in] uhbt: The summed volume flux through zonal faces[in] uhbt_aux: A second set of summed volume fluxes through zonal faces [H m2 s-1 ~> m3 s-1 or kg s-1].[out] u_cor: The zonal velocitiess (u with a barotropic correction) that give uhbt as the depth-integrated transport, m s-1.[out] u_cor_aux: The zonal velocities (u with a barotropic correction) that give uhbt_aux as the depth-integrated transports [m s-1].bt_cont: A structure with elements that describe the effective open face areas as a function of barotropic flow.
-
subroutine
zonal_flux_layer(u u, h h, h_L h_L, h_R h_R, uh uh, duhdu duhdu, visc_rem visc_rem, dt dt, G G, j j, ish ish, ieh ieh, do_I do_I, vol_CFL vol_CFL, OBC OBC)¶ Evaluates the zonal mass or volume fluxes in a layer.
- Parameters
[inout] g: Ocean’s grid structure.[in] u: Zonal velocity [m s-1].[in] visc_rem: Both the fraction of the momentum originally in a layer that remains after a time-step of viscosity, and the fraction of a time-step’s worth of a barotropic acceleration that a layer experiences after viscosity is applied. Non-dimensional between 0 (at the bottom) and 1 (far above the bottom).[in] h: Layer thickness [H ~> m or kg m-2].[in] h_l: Left thickness [H ~> m or kg m-2].[in] h_r: Right thickness [H ~> m or kg m-2].[inout] uh: Zonal mass or volume transport [H m2 s-1 ~> m3 s-1 or kg s-1].[inout] duhdu: Partial derivative of uh with u [H m ~> m2 or kg m-1].[in] dt: Time increment [s].[in] j: Spatial index.[in] ish: Start of index range.[in] ieh: End of index range.[in] do_i: Which i values to work on.[in] vol_cfl: If true, rescale the ratio of face areas to the cell areas when estimating the CFL number.obc: Open boundaries control structure.
-
subroutine
zonal_face_thickness(u u, h h, h_L h_L, h_R h_R, h_u h_u, dt dt, G G, LB LB, vol_CFL vol_CFL, marginal marginal, visc_rem_u visc_rem_u, OBC OBC)¶ Sets the effective interface thickness at each zonal velocity point.
- Parameters
[inout] g: Ocean’s grid structure.[in] u: Zonal velocity [m s-1].[in] h: Layer thickness used to calculate fluxes [H ~> m or kg m-2].[in] h_l: Left thickness in the reconstruction [H ~> m or kg m-2].[in] h_r: Right thickness in the reconstruction [H ~> m or kg m-2].[inout] h_u: Thickness at zonal faces [H ~> m or kg m-2].[in] dt: Time increment [s].[in] lb: Loop bounds structure.[in] vol_cfl: If true, rescale the ratio of face areas to the cell areas when estimating the CFL number.[in] marginal: If true, report the marginal face thicknesses; otherwise report transport-averaged thicknesses.[in] visc_rem_u: Both the fraction of the momentum originally in a layer that remains after a time-step of viscosity, and the fraction of a time-step’s worth of a barotropic acceleration that a layer experiences after viscosity is applied. Non-dimensional between 0 (at the bottom) and 1 (far above the bottom).obc: Open boundaries control structure.
-
subroutine
zonal_flux_adjust(u u, h_in h_in, h_L h_L, h_R h_R, uhbt uhbt, uh_tot_0 uh_tot_0, duhdu_tot_0 duhdu_tot_0, du du, du_max_CFL du_max_CFL, du_min_CFL du_min_CFL, dt dt, G G, CS CS, visc_rem visc_rem, j j, ish ish, ieh ieh, do_I_in do_I_in, full_precision full_precision, uh_3d uh_3d, OBC OBC)¶ Returns the barotropic velocity adjustment that gives the desired barotropic (layer-summed) transport.
- Parameters
[inout] g: Ocean’s grid structure.[in] u: Zonal velocity [m s-1].[in] h_in: Layer thickness used to calculate fluxes [H ~> m or kg m-2].[in] h_l: Left thickness in the reconstruction [H ~> m or kg m-2].[in] h_r: Right thickness in the reconstruction [H ~> m or kg m-2].[in] visc_rem: Both the fraction of the momentum originally in a layer that remains after a time-step of viscosity, and the fraction of a time-step’s worth of a barotropic acceleration that a layer experiences after viscosity is applied. Non-dimensional between 0 (at the bottom) and 1 (far above the bottom).[in] uhbt: The summed volume flux through zonal faces [H m2 s-1 ~> m3 s-1 or kg s-1].[in] du_max_cfl: Maximum acceptable value of du [m s-1].[in] du_min_cfl: Minimum acceptable value of du [m s-1].[in] uh_tot_0: The summed transport with 0 adjustment [H m2 s-1 ~> m3 s-1 or kg s-1].[in] duhdu_tot_0: The partial derivative of du_err with du at 0 adjustment [H m ~> m2 or kg m-1].[out] du: The barotropic velocity adjustment [m s-1].[in] dt: Time increment [s].cs: This module’s control structure.[in] j: Spatial index.[in] ish: Start of index range.[in] ieh: End of index range.[in] do_i_in: A logical flag indicating which I values to work on.[in] full_precision: A flag indicating how carefully to iterate. The default is .true. (more accurate).[inout] uh_3d: Volume flux through zonal faces = u*h*dy [H m2 s-1 ~> m3 s-1 or kg s-1].obc: Open boundaries control structure.
-
subroutine
set_zonal_bt_cont(u u, h_in h_in, h_L h_L, h_R h_R, BT_cont BT_cont, uh_tot_0 uh_tot_0, duhdu_tot_0 duhdu_tot_0, du_max_CFL du_max_CFL, du_min_CFL du_min_CFL, dt dt, G G, US US, CS CS, visc_rem visc_rem, visc_rem_max visc_rem_max, j j, ish ish, ieh ieh, do_I do_I)¶ Sets a structure that describes the zonal barotropic volume or mass fluxes as a function of barotropic flow to agree closely with the sum of the layer’s transports.
- Parameters
[inout] g: Ocean’s grid structure.[in] u: Zonal velocity [m s-1].[in] h_in: Layer thickness used to calculate fluxes [H ~> m or kg m-2].[in] h_l: Left thickness in the reconstruction [H ~> m or kg m-2].[in] h_r: Right thickness in the reconstruction [H ~> m or kg m-2].[inout] bt_cont: A structure with elements that describe the effective open face areas as a function of barotropic flow.[in] uh_tot_0: The summed transport with 0 adjustment [H m2 s-1 ~> m3 s-1 or kg s-1].[in] duhdu_tot_0: The partial derivative of du_err with du at 0 adjustment [H m ~> m2 or kg m-1].[in] du_max_cfl: Maximum acceptable value of du [m s-1].[in] du_min_cfl: Minimum acceptable value of du [m s-1].[in] dt: Time increment [s].[in] us: A dimensional unit scaling typecs: This module’s control structure.[in] visc_rem: Both the fraction of the momentum originally in a layer that remains after a time-step of viscosity, and the fraction of a time-step’s worth of a barotropic acceleration that a layer experiences after viscosity is applied. Non-dimensional between 0 (at the bottom) and 1 (far above the bottom).[in] visc_rem_max: Maximum allowable visc_rem.[in] j: Spatial index.[in] ish: Start of index range.[in] ieh: End of index range.[in] do_i: A logical flag indicating which I values to work on.
-
subroutine
meridional_mass_flux(v v, h_in h_in, vh vh, dt dt, G G, GV GV, US US, CS CS, LB LB, vhbt vhbt, OBC OBC, visc_rem_v visc_rem_v, v_cor v_cor, vhbt_aux vhbt_aux, v_cor_aux v_cor_aux, BT_cont BT_cont)¶ Calculates the mass or volume fluxes through the meridional faces, and other related quantities.
- Parameters
[inout] g: Ocean’s grid structure.[in] gv: Ocean’s vertical grid structure.[in] v: Meridional velocity [m s-1].[in] h_in: Layer thickness used to calculate fluxes [H ~> m or kg m-2].[out] vh: Volume flux through meridional faces = v*h*dx [H m2 s-1 ~> m3 s-1 or kg s-1].[in] dt: Time increment [s].[in] us: A dimensional unit scaling typecs: This module’s control structure.[in] lb: Loop bounds structure.obc: Open boundary condition type specifies whether, where, and what open boundary conditions are used.[in] visc_rem_v: Both the fraction of the momentum[in] vhbt: The summed volume flux through meridional faces [H m2 s-1 ~> m3 s-1 or kg s-1].[in] vhbt_aux: A second set of summed volume fluxes through meridional faces [H m2 s-1 ~> m3 s-1 or kg s-1].[out] v_cor: The meridional velocitiess (v with a barotropic correction) that give vhbt as the depth-integrated transport [m s-1].[out] v_cor_aux: The meridional velocities (v with a barotropic correction) that give vhbt_aux as the depth-integrated transports [m s-1].bt_cont: A structure with elements that describe the effective open face areas as a function of barotropic flow.
-
subroutine
merid_flux_layer(v v, h h, h_L h_L, h_R h_R, vh vh, dvhdv dvhdv, visc_rem visc_rem, dt dt, G G, J J, ish ish, ieh ieh, do_I do_I, vol_CFL vol_CFL, OBC OBC)¶ Evaluates the meridional mass or volume fluxes in a layer.
- Parameters
[inout] g: Ocean’s grid structure.[in] v: Meridional velocity [m s-1].[in] visc_rem: Both the fraction of the momentum originally in a layer that remains after a time-step of viscosity, and the fraction of a time-step’s worth of a barotropic acceleration that a layer experiences after viscosity is applied. Non-dimensional between 0 (at the bottom) and 1 (far above the bottom).[in] h: Layer thickness used to calculate fluxes, [H ~> m or kg m-2].[in] h_l: Left thickness in the reconstruction [H ~> m or kg m-2].[in] h_r: Right thickness in the reconstruction [H ~> m or kg m-2].[inout] vh: Meridional mass or volume transport [H m2 s-1 ~> m3 s-1 or kg s-1].[inout] dvhdv: Partial derivative of vh with v [H m ~> m2 or kg m-1].[in] dt: Time increment [s].[in] j: Spatial index.[in] ish: Start of index range.[in] ieh: End of index range.[in] do_i: Which i values to work on.[in] vol_cfl: If true, rescale the ratio of face areas to the cell areas when estimating the CFL number.obc: Open boundaries control structure.
-
subroutine
merid_face_thickness(v v, h h, h_L h_L, h_R h_R, h_v h_v, dt dt, G G, LB LB, vol_CFL vol_CFL, marginal marginal, visc_rem_v visc_rem_v, OBC OBC)¶ Sets the effective interface thickness at each meridional velocity point.
- Parameters
[inout] g: Ocean’s grid structure.[in] v: Meridional velocity [m s-1].[in] h: Layer thickness used to calculate fluxes, [H ~> m or kg m-2].[in] h_l: Left thickness in the reconstruction, [H ~> m or kg m-2].[in] h_r: Right thickness in the reconstruction, [H ~> m or kg m-2].[inout] h_v: Thickness at meridional faces, [H ~> m or kg m-2].[in] dt: Time increment [s].[in] lb: Loop bounds structure.[in] vol_cfl: If true, rescale the ratio of face areas to the cell areas when estimating the CFL number.[in] marginal: If true, report the marginal face thicknesses; otherwise report transport-averaged thicknesses.[in] visc_rem_v: Both the fraction of the momentum originally in a layer that remains after a time-step of viscosity, and the fraction of a time-step’s worth of a barotropic acceleration that a layer experiences after viscosity is applied. Non-dimensional between 0 (at the bottom) and 1 (far above the bottom).obc: Open boundaries control structure.
-
subroutine
meridional_flux_adjust(v v, h_in h_in, h_L h_L, h_R h_R, vhbt vhbt, vh_tot_0 vh_tot_0, dvhdv_tot_0 dvhdv_tot_0, dv dv, dv_max_CFL dv_max_CFL, dv_min_CFL dv_min_CFL, dt dt, G G, CS CS, visc_rem visc_rem, j j, ish ish, ieh ieh, do_I_in do_I_in, full_precision full_precision, vh_3d vh_3d, OBC OBC)¶ Returns the barotropic velocity adjustment that gives the desired barotropic (layer-summed) transport.
- Parameters
[inout] g: Ocean’s grid structure.[in] v: Meridional velocity [m s-1].[in] h_in: Layer thickness used to calculate fluxes [H ~> m or kg m-2].[in] h_l: Left thickness in the reconstruction [H ~> m or kg m-2].[in] h_r: Right thickness in the reconstruction [H ~> m or kg m-2].[in] visc_rem: Both the fraction of the momentum originally in a layer that remains after a time-step of viscosity, and the fraction of a time-step’s worth of a barotropic acceleration that a layer experiences after viscosity is applied. Non-dimensional between 0 (at the bottom) and 1 (far above the bottom).[in] vhbt: The summed volume flux through meridional faces[in] dv_max_cfl: Maximum acceptable value of dv [m s-1].[in] dv_min_cfl: Minimum acceptable value of dv [m s-1].[in] vh_tot_0: The summed transport with 0 adjustment [H m2 s-1 ~> m3 s-1 or kg s-1].[in] dvhdv_tot_0: The partial derivative of dv_err with dv at 0 adjustment [H m ~> m2 or kg m-1].[out] dv: The barotropic velocity adjustment [m s-1].[in] dt: Time increment [s].cs: This module’s control structure.[in] j: Spatial index.[in] ish: Start of index range.[in] ieh: End of index range.[in] do_i_in: A flag indicating which I values to work on.[in] full_precision: A flag indicating how carefully to iterate. The default is .true. (more accurate).[inout] vh_3d: Volume flux through meridionalobc: Open boundaries control structure.
-
subroutine
set_merid_bt_cont(v v, h_in h_in, h_L h_L, h_R h_R, BT_cont BT_cont, vh_tot_0 vh_tot_0, dvhdv_tot_0 dvhdv_tot_0, dv_max_CFL dv_max_CFL, dv_min_CFL dv_min_CFL, dt dt, G G, US US, CS CS, visc_rem visc_rem, visc_rem_max visc_rem_max, j j, ish ish, ieh ieh, do_I do_I)¶ Sets of a structure that describes the meridional barotropic volume or mass fluxes as a function of barotropic flow to agree closely with the sum of the layer’s transports.
- Parameters
[inout] g: Ocean’s grid structure.[in] v: Meridional velocity [m s-1].[in] h_in: Layer thickness used to calculate fluxes, [H ~> m or kg m-2].[in] h_l: Left thickness in the reconstruction, [H ~> m or kg m-2].[in] h_r: Right thickness in the reconstruction, [H ~> m or kg m-2].[inout] bt_cont: A structure with elements that describe the effective open face areas as a function of barotropic flow.[in] vh_tot_0: The summed transport with 0 adjustment [H m2 s-1 ~> m3 s-1 or kg s-1].[in] dvhdv_tot_0: The partial derivative of du_err with dv at 0 adjustment [H m ~> m2 or kg m-1].[in] dv_max_cfl: Maximum acceptable value of dv [m s-1].[in] dv_min_cfl: Minimum acceptable value of dv [m s-1].[in] dt: Time increment [s].[in] us: A dimensional unit scaling typecs: This module’s control structure.[in] visc_rem: Both the fraction of the momentum originally in a layer that remains after a time-step of viscosity, and the fraction of a time-step’s worth of a barotropic acceleration that a layer experiences after viscosity is applied. Non-dimensional between 0 (at the bottom) and 1 (far above the bottom).[in] visc_rem_max: Maximum allowable visc_rem.[in] j: Spatial index.[in] ish: Start of index range.[in] ieh: End of index range.[in] do_i: A logical flag indicating which I values to work on.
-
subroutine
ppm_reconstruction_x(h_in h_in, h_L h_L, h_R h_R, G G, LB LB, h_min h_min, monotonic monotonic, simple_2nd simple_2nd, OBC OBC)¶ Calculates left/right edge values for PPM reconstruction.
- Parameters
[in] g: Ocean’s grid structure.[in] h_in: Layer thickness [H ~> m or kg m-2].[out] h_l: Left thickness in the reconstruction, [H ~> m or kg m-2].[out] h_r: Right thickness in the reconstruction, [H ~> m or kg m-2].[in] lb: Active loop bounds structure.[in] h_min: The minimum thickness that can be obtained by a concave parabolic fit.[in] monotonic: If true, use the Colella & Woodward monotonic limiter. Otherwise use a simple positive-definite limiter.[in] simple_2nd: If true, use the arithmetic mean thicknesses as the default edge values for a simple 2nd order scheme.obc: Open boundaries control structure.
-
subroutine
ppm_reconstruction_y(h_in h_in, h_L h_L, h_R h_R, G G, LB LB, h_min h_min, monotonic monotonic, simple_2nd simple_2nd, OBC OBC)¶ Calculates left/right edge values for PPM reconstruction.
- Parameters
[in] g: Ocean’s grid structure.[in] h_in: Layer thickness [H ~> m or kg m-2].[out] h_l: Left thickness in the reconstruction, [H ~> m or kg m-2].[out] h_r: Right thickness in the reconstruction, [H ~> m or kg m-2].[in] lb: Active loop bounds structure.[in] h_min: The minimum thickness that can be obtained by a concave parabolic fit.[in] monotonic: If true, use the Colella & Woodward monotonic limiter. Otherwise use a simple positive-definite limiter.[in] simple_2nd: If true, use the arithmetic mean thicknesses as the default edge values for a simple 2nd order scheme.obc: Open boundaries control structure.
-
subroutine
ppm_limit_pos(h_in h_in, h_L h_L, h_R h_R, h_min h_min, G G, iis iis, iie iie, jis jis, jie jie)¶ This subroutine limits the left/right edge values of the PPM reconstruction to give a reconstruction that is positive-definite. Here this is reinterpreted as giving a constant thickness if the mean thickness is less than h_min, with a minimum of h_min otherwise.
- Parameters
[in] g: Ocean’s grid structure.[in] h_in: Layer thickness [H ~> m or kg m-2].[inout] h_l: Left thickness in the reconstruction [H ~> m or kg m-2].[inout] h_r: Right thickness in the reconstruction [H ~> m or kg m-2].[in] h_min: The minimum thickness that can be obtained by a concave parabolic fit.[in] iis: Start of i index range.[in] iie: End of i index range.[in] jis: Start of j index range.[in] jie: End of j index range.
-
subroutine
ppm_limit_cw84(h_in h_in, h_L h_L, h_R h_R, G G, iis iis, iie iie, jis jis, jie jie)¶ This subroutine limits the left/right edge values of the PPM reconstruction according to the monotonic prescription of Colella and Woodward, 1984.
- Parameters
[in] g: Ocean’s grid structure.[in] h_in: Layer thickness [H ~> m or kg m-2].[inout] h_l: Left thickness in the reconstruction, [H ~> m or kg m-2].[inout] h_r: Right thickness in the reconstruction, [H ~> m or kg m-2].[in] iis: Start of i index range.[in] iie: End of i index range.[in] jis: Start of j index range.[in] jie: End of j index range.
-
real function mom_continuity_ppm::ratio_max(a a, b b, maxrat maxrat) Return the maximum ratio of a/b or maxrat.
- Return
Return value.
- Parameters
[in] a: Numerator[in] b: Denominator[in] maxrat: Maximum value of ratio.
-
subroutine, public mom_continuity_ppm::continuity_ppm_init(Time Time, G G, GV GV, param_file param_file, diag diag, CS CS) Initializes continuity_ppm_cs.
- Parameters
[in] time: Time increment [s].[in] g: The ocean’s grid structure.[in] gv: Vertical grid structure.[in] param_file: A structure indicating the open file to parse for model parameter values.[inout] diag: A structure that is used to regulate diagnostic output.cs: Module’s control structure.
-
integer function, public mom_continuity_ppm::continuity_ppm_stencil(CS CS) continuity_PPM_stencil returns the continuity solver stencil size
- Return
The continuity solver stencil size with the current settings.
- Parameters
cs: Module’s control structure.
-
subroutine, public mom_continuity_ppm::continuity_ppm_end(CS CS) Destructor for continuity_ppm_cs.
- Parameters
cs: Module’s control structure.
-
integer
-
namespace
mom_controlled_forcing¶ Use control-theory to adjust the surface heat flux and precipitation.
Adjustments are based on the time-mean or periodically (seasonally) varying anomalies from the observed state.
The techniques behind this are described in Hallberg and Adcroft (2018, in prep.).
By Robert Hallberg, July 2011 *
This program contains the subroutines that use control-theory * to adjust the surface heat flux and precipitation, based on the * time-mean or periodically (seasonally) varying anomalies from the * observed state. The techniques behind this are described in * Hallberg and Adcroft (2011, in prep.). *
Macros written all in capital letters are defined in MOM_memory.h. *
A small fragment of the grid is shown below: *
j+1 x ^ x ^ x At x: q * j+1 > o > o > At ^: v, tauy * j x ^ x ^ x At >: u, taux * j > o > o > At o: h, fluxes. * j-1 x ^ x ^ x * i-1 i i+1 At x & ^: * i i+1 At > & o: *
The boundaries always run through q grid points (x). *
Unnamed Group
-
subroutine, public mom_controlled_forcing::apply_ctrl_forcing(SST_anom SST_anom, SSS_anom SSS_anom, SSS_mean SSS_mean, virt_heat virt_heat, virt_precip virt_precip, day_start day_start, dt dt, G G, CS CS) This subroutine calls any of the other subroutines in this file that are needed to specify the current surface forcing fields.
- Parameters
[inout] g: The ocean’s grid structure.[in] sst_anom: The sea surface temperature anomalies [degC].[in] sss_anom: The sea surface salinity anomlies [ppt].[in] sss_mean: The mean sea surface salinity [ppt].[inout] virt_heat: Virtual (corrective) heat fluxes that are augmented in this subroutine [W m-2].[inout] virt_precip: Virtual (corrective) precipitation fluxes that are augmented in this subroutine [kg m-2 s-1].[in] day_start: Start time of the fluxes.[in] dt: Length of time over which these fluxes will be applied [s].cs: A pointer to the control structure returned by a previous call to ctrl_forcing_init.
-
integer function mom_controlled_forcing::periodic_int(rval rval, num_period num_period) This function maps rval into an integer in the range from 1 to num_period.
- Return
Return value.
- Parameters
[in] rval: Input for mapping.[in] num_period: Maximum output.
-
real function mom_controlled_forcing::periodic_real(rval rval, num_period num_period) This function shifts rval by an integer multiple of num_period so that 0 <= val_out < num_period.
- Return
Return value.
- Parameters
[in] rval: Input to be shifted into valid range.[in] num_period: Maximum valid value.
-
subroutine, public mom_controlled_forcing::register_ctrl_forcing_restarts(G G, param_file param_file, CS CS, restart_CS restart_CS) This subroutine is used to allocate and register any fields in this module that should be written to or read from the restart file.
- Parameters
[in] g: The ocean’s grid structure.[in] param_file: A structure indicating the open file to parse for model parameter values.cs: A pointer that is set to point to the control structure for this module.restart_cs: A pointer to the restart control structure.
-
subroutine, public mom_controlled_forcing::controlled_forcing_init(Time Time, G G, param_file param_file, diag diag, CS CS) Set up this modules control structure.
- Parameters
[in] time: The current model time.[in] g: The ocean’s grid structure.[in] param_file: A structure indicating the open file to parse for model parameter values.[in] diag: A structure that is used to regulate diagnostic output.cs: A pointer that is set to point to the control structure for this module.
-
subroutine, public mom_controlled_forcing::controlled_forcing_end(CS CS) Clean up this modules control structure.
- Parameters
cs: A pointer to the control structure returned by a previous call to controlled_forcing_init, it will be deallocated here.
-
namespace
mom_coord_initialization¶ Initializes fixed aspects of the related to its vertical coordinate.
Functions
-
subroutine, public mom_coord_initialization::mom_initialize_coord(GV GV, US US, PF PF, write_geom write_geom, output_dir output_dir, tv tv, max_depth max_depth) MOM_initialize_coord sets up time-invariant quantities related to MOM6’s vertical coordinate.
- Parameters
[inout] gv: Ocean vertical grid structure.[in] us: A dimensional unit scaling type[in] pf: A structure indicating the open file to parse for model parameter values.[in] write_geom: If true, write grid geometry files.[in] output_dir: The directory into which to write files.[inout] tv: The thermodynamic variable structure.[in] max_depth: The ocean’s maximum depth [Z ~> m].
-
subroutine
set_coord_from_gprime(Rlay Rlay, g_prime g_prime, GV GV, US US, param_file param_file)¶ Sets the layer densities (Rlay) and the interface reduced gravities (g).
- Parameters
[out] rlay: The layers’ target coordinate values (potential density) [kg m-3].[out] g_prime: The reduced gravity across the interfaces [L2 Z-1 T-2 ~> m s-2].[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters
-
subroutine
set_coord_from_layer_density(Rlay Rlay, g_prime g_prime, GV GV, US US, param_file param_file)¶ Sets the layer densities (Rlay) and the interface reduced gravities (g).
- Parameters
[out] rlay: The layers’ target coordinate values (potential density) [kg m-3].[out] g_prime: The reduced gravity across the interfaces [L2 Z-1 T-2 ~> m s-2].[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters
-
subroutine
set_coord_from_ts_ref(Rlay Rlay, g_prime g_prime, GV GV, US US, param_file param_file, eqn_of_state eqn_of_state, P_Ref P_Ref)¶ Sets the layer densities (Rlay) and the interface reduced gravities (g) from a profile of g’.
- Parameters
[out] rlay: The layers’ target coordinate values (potential density) [kg m-3].[out] g_prime: The reduced gravity across the interfaces [L2 Z-1 T-2 ~> m s-2].[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameterseqn_of_state: integer selecting the equation of state.[in] p_ref: The coordinate-density reference pressure [Pa].
-
subroutine
set_coord_from_ts_profile(Rlay Rlay, g_prime g_prime, GV GV, US US, param_file param_file, eqn_of_state eqn_of_state, P_Ref P_Ref)¶ Sets the layer densities (Rlay) and the interface reduced gravities (g) from a T-S profile.
- Parameters
[out] rlay: The layers’ target coordinate values (potential density) [kg m-3].[out] g_prime: The reduced gravity across the interfaces [L2 Z-1 T-2 ~> m s-2].[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameterseqn_of_state: integer that selects equation of state.[in] p_ref: The coordinate-density reference pressure [Pa].
-
subroutine
set_coord_from_ts_range(Rlay Rlay, g_prime g_prime, GV GV, US US, param_file param_file, eqn_of_state eqn_of_state, P_Ref P_Ref)¶ Sets the layer densities (Rlay) and the interface reduced gravities (g) from a linear T-S profile.
- Parameters
[out] rlay: The layers’ target coordinate values (potential density) [kg m-3].[out] g_prime: The reduced gravity across the interfaces [L2 Z-1 T-2 ~> m s-2].[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameterseqn_of_state: integer that selects equation of state[in] p_ref: The coordinate-density reference pressure [Pa]
-
subroutine
set_coord_from_file(Rlay Rlay, g_prime g_prime, GV GV, US US, param_file param_file)¶ - Parameters
[out] rlay: The layers’ target coordinate values (potential density) [kg m-3].[out] g_prime: The reduced gravity across the interfaces [L2 Z-1 T-2 ~> m s-2].[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters
-
subroutine
set_coord_linear(Rlay Rlay, g_prime g_prime, GV GV, US US, param_file param_file)¶ Sets the layer densities (Rlay) and the interface reduced gravities (g) according to a linear profile starting at a reference surface layer density and spanning a range of densities to the bottom defined by the parameter RLAY_RANGE (defaulting to 2.0 if not defined)
- Parameters
[out] rlay: The layers’ target coordinate values (potential density) [kg m-3].[out] g_prime: The reduced gravity across the interfaces [L2 Z-1 T-2 ~> m s-2].[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters
-
subroutine
set_coord_to_none(Rlay Rlay, g_prime g_prime, GV GV, US US, param_file param_file)¶ Sets Rlay to Rho0 and g_prime to zero except for the free surface. This is for use only in ALE mode where Rlay should not be used and g_prime(1) alone might be used.
- Parameters
[out] rlay: The layers’ target coordinate values (potential density) [kg m-3].[out] g_prime: The reduced gravity across the interfaces, [L2 Z-1 T-2 ~> m s-2].[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters
-
subroutine
write_vertgrid_file(GV GV, US US, param_file param_file, directory directory)¶ Writes out a file containing any available data related to the vertical grid used by the MOM ocean model.
- Parameters
[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters[in] directory: The directory into which to place the file.
Variables
-
character(len=40) mom_coord_initialization::mdl = "MOM_coord_initialization" This module’s name.
-
-
namespace
mom_coriolisadv¶ Accelerations due to the Coriolis force and momentum advection.
This file contains the subroutine that calculates the time derivatives of the velocities due to Coriolis acceleration and momentum advection. This subroutine uses either a vorticity advection scheme from Arakawa and Hsu, Mon. Wea. Rev. 1990, or Sadourny’s (JAS 1975) energy conserving scheme. Both have been modified to use general orthogonal coordinates as described in Arakawa and Lamb, Mon. Wea. Rev. 1981. Both schemes are second order accurate, and allow for vanishingly small layer thicknesses. The Arakawa and Hsu scheme globally conserves both total energy and potential enstrophy in the limit of nondivergent flow. Sadourny’s energy conserving scheme conserves energy if the flow is nondivergent or centered difference thickness fluxes are used.
Two sets of boundary conditions have been coded in the definition of relative vorticity. These are written as: NOSLIP defined (in spherical coordinates): relvort = dv/dx (east & west), with v = 0. relvort = -sec(Q) * d(u cos(Q))/dy (north & south), with u = 0.
NOSLIP not defined (free slip): relvort = 0 (all boundaries)
with Q temporarily defined as latitude. The free slip boundary condition is much more natural on a C-grid.
A small fragment of the grid is shown below:
j+1 x ^ x ^ x At x: q, CoriolisBu j+1 > o > o > At ^: v, CAv, vh j x ^ x ^ x At >: u, CAu, uh, a, b, c, d j > o > o > At o: h, KE j-1 x ^ x ^ x i-1 i i+1 At x & ^: i i+1 At > & o:
The boundaries always run through q grid points (x).
Unnamed Group
-
integer, parameter mom_coriolisadv::sadourny75_energy = 1 Enumeration values for Coriolis_Scheme.
-
integer, parameter mom_coriolisadv::arakawa_hsu90 = 2 Enumeration values for Coriolis_Scheme.
-
integer, parameter mom_coriolisadv::robust_enstro = 3 Enumeration values for Coriolis_Scheme.
-
integer, parameter mom_coriolisadv::sadourny75_enstro = 4 Enumeration values for Coriolis_Scheme.
-
integer, parameter mom_coriolisadv::arakawa_lamb81 = 5 Enumeration values for Coriolis_Scheme.
-
integer, parameter mom_coriolisadv::al_blend = 6 Enumeration values for Coriolis_Scheme.
-
character*(20), parameter mom_coriolisadv::sadourny75_energy_string = "SADOURNY75_ENERGY" Enumeration values for Coriolis_Scheme.
-
character*(20), parameter mom_coriolisadv::arakawa_hsu_string = "ARAKAWA_HSU90" Enumeration values for Coriolis_Scheme.
-
character*(20), parameter mom_coriolisadv::robust_enstro_string = "ROBUST_ENSTRO" Enumeration values for Coriolis_Scheme.
-
character*(20), parameter mom_coriolisadv::sadourny75_enstro_string = "SADOURNY75_ENSTRO" Enumeration values for Coriolis_Scheme.
-
character*(20), parameter mom_coriolisadv::arakawa_lamb_string = "ARAKAWA_LAMB81" Enumeration values for Coriolis_Scheme.
-
character*(20), parameter mom_coriolisadv::al_blend_string = "ARAKAWA_LAMB_BLEND" Enumeration values for Coriolis_Scheme.
-
integer, parameter mom_coriolisadv::ke_arakawa = 10 Enumeration values for KE_Scheme.
-
integer, parameter mom_coriolisadv::ke_simple_gudonov = 11 Enumeration values for Coriolis_Scheme.
-
integer, parameter mom_coriolisadv::ke_gudonov = 12 Enumeration values for Coriolis_Scheme.
-
character*(20), parameter mom_coriolisadv::ke_arakawa_string = "KE_ARAKAWA" Enumeration values for Coriolis_Scheme.
-
character*(20), parameter mom_coriolisadv::ke_simple_gudonov_string = "KE_SIMPLE_GUDONOV" Enumeration values for Coriolis_Scheme.
-
character*(20), parameter mom_coriolisadv::ke_gudonov_string = "KE_GUDONOV" Enumeration values for Coriolis_Scheme.
-
integer, parameter mom_coriolisadv::pv_adv_centered = 21 Enumeration values for PV_Adv_Scheme.
-
integer, parameter mom_coriolisadv::pv_adv_upwind1 = 22 Enumeration values for Coriolis_Scheme.
-
character*(20), parameter mom_coriolisadv::pv_adv_centered_string = "PV_ADV_CENTERED" Enumeration values for Coriolis_Scheme.
-
character*(20), parameter mom_coriolisadv::pv_adv_upwind1_string = "PV_ADV_UPWIND1" Enumeration values for Coriolis_Scheme.
-
subroutine, public mom_coriolisadv::coradcalc(u u, v v, h h, uh uh, vh vh, CAu CAu, CAv CAv, OBC OBC, AD AD, G G, GV GV, US US, CS CS) Calculates the Coriolis and momentum advection contributions to the acceleration.
- Parameters
[in] g: Ocen grid structure[in] gv: Vertical grid structure[in] u: Zonal velocity [m s-1][in] v: Meridional velocity [m s-1][in] h: Layer thickness [H ~> m or kg m-2][in] uh: Zonal transport u*h*dy [H m2 s-1 ~> m3 s-1 or kg s-1][in] vh: Meridional transport v*h*dx [H m2 s-1 ~> m3 s-1 or kg s-1][out] cau: Zonal acceleration due to Coriolis and momentum advection [m s-2].[out] cav: Meridional acceleration due to Coriolis and momentum advection [m s-2].obc: Open boundary control structure[inout] ad: Storage for acceleration diagnostics[in] us: A dimensional unit scaling typecs: Control structure for MOM_CoriolisAdv
-
subroutine
gradke(u u, v v, h h, KE KE, KEx KEx, KEy KEy, k k, OBC OBC, G G, CS CS)¶ Calculates the acceleration due to the gradient of kinetic energy.
- Parameters
[in] g: Ocen grid structure[in] u: Zonal velocity [m s-1][in] v: Meridional velocity [m s-1][in] h: Layer thickness [H ~> m or kg m-2][out] ke: Kinetic energy [m2 s-2][out] kex: Zonal acceleration due to kinetic energy gradient [m s-2][out] key: Meridional acceleration due to kinetic energy gradient [m s-2][in] k: Layer number to calculate forobc: Open boundary control structurecs: Control structure for MOM_CoriolisAdv
-
subroutine, public mom_coriolisadv::coriolisadv_init(Time Time, G G, param_file param_file, diag diag, AD AD, CS CS) Initializes the control structure for coriolisadv_cs.
- Parameters
[in] time: Current model time[in] g: Ocean grid structure[in] param_file: Runtime parameter handles[inout] diag: Diagnostics control structure[inout] ad: Strorage for acceleration diagnosticscs: Control structure fro MOM_CoriolisAdv
-
subroutine, public mom_coriolisadv::coriolisadv_end(CS CS) Destructor for coriolisadv_cs.
- Parameters
cs: Control structure fro MOM_CoriolisAdv
-
-
namespace
mom_cpu_clock¶ Wraps the MPP cpu clock functions.
-
namespace
mom_cvmix_conv¶ Interface to CVMix convection scheme.
Unnamed Group
-
character(len=40) mom_cvmix_conv::mdl = "MOM_CVMix_conv" This module’s name.
-
logical function, public mom_cvmix_conv::cvmix_conv_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS) Initialized the CVMix convection mixing routine.
- Parameters
[in] time: The current time.[in] g: Grid structure.[in] gv: Vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: Run-time parameter file handle[inout] diag: Diagnostics control structure.cs: This module’s control structure.
-
subroutine, public mom_cvmix_conv::calculate_cvmix_conv(h h, tv tv, G G, GV GV, US US, CS CS, hbl hbl) Subroutine for calculating enhanced diffusivity/viscosity due to convection via CVMix.
- Parameters
[in] g: Grid structure.[in] gv: Vertical grid structure.[in] us: A dimensional unit scaling type[in] h: Layer thickness [H ~> m or kg m-2].[in] tv: Thermodynamics structure.cs: The control structure returned by a previous call to CVMix_conv_init.hbl: Depth of ocean boundary layer [m]
-
logical function, public mom_cvmix_conv::cvmix_conv_is_used(param_file param_file) Reads the parameter “USE_CVMix_CONVECTION” and returns state. This function allows other modules to know whether this parameterization will be used without needing to duplicate the log entry.
- Parameters
[in] param_file: A structure to parse for run-time parameters
-
subroutine, public mom_cvmix_conv::cvmix_conv_end(CS CS) Clear pointers and dealocate memory.
- Parameters
cs: Control structure for this module that will be deallocated in this subroutine
-
-
namespace
mom_cvmix_ddiff¶ Interface to CVMix double diffusion scheme.
Unnamed Group
-
character(len=40) mom_cvmix_ddiff::mdl = "MOM_CVMix_ddiff" This module’s name.
-
logical function, public mom_cvmix_ddiff::cvmix_ddiff_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS) Initialized the CVMix double diffusion module.
- Parameters
[in] time: The current time.[in] g: Grid structure.[in] gv: Vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: Run-time parameter file handle[inout] diag: Diagnostics control structure.cs: This module’s control structure.
-
subroutine, public mom_cvmix_ddiff::compute_ddiff_coeffs(h h, tv tv, G G, GV GV, US US, j j, Kd_T Kd_T, Kd_S Kd_S, CS CS) Subroutine for computing vertical diffusion coefficients for the double diffusion mixing parameterization.
- Parameters
[in] g: Grid structure.[in] gv: Vertical grid structure.[in] us: A dimensional unit scaling type[in] h: Layer thickness [H ~> m or kg m-2].[in] tv: Thermodynamics structure.[out] kd_t: Interface double diffusion diapycnal diffusivity for temp [Z2 T-1 ~> m2 s-1].[out] kd_s: Interface double diffusion diapycnal diffusivity for salt [Z2 T-1 ~> m2 s-1].cs: The control structure returned by a previous call to CVMix_ddiff_init.[in] j: Meridional grid indice.
-
logical function, public mom_cvmix_ddiff::cvmix_ddiff_is_used(param_file param_file) Reads the parameter “USE_CVMIX_DDIFF” and returns state. This function allows other modules to know whether this parameterization will be used without needing to duplicate the log entry.
- Parameters
[in] param_file: A structure to parse for run-time parameters
-
subroutine, public mom_cvmix_ddiff::cvmix_ddiff_end(CS CS) Clear pointers and dealocate memory.
- Parameters
cs: Control structure for this module that will be deallocated in this subroutine
-
-
namespace
mom_cvmix_kpp¶ Provides the K-Profile Parameterization (KPP) of Large et al., 1994, via CVMix.
Unnamed Group
-
logical function, public mom_cvmix_kpp::kpp_init(paramFile paramFile, G G, GV GV, US US, diag diag, Time Time, CS CS, passive passive, Waves Waves) Initialize the CVMix KPP module and set up diagnostics Returns True if KPP is to be used, False otherwise.
- Parameters
[in] paramfile: File parser[in] g: Ocean grid[in] gv: Vertical grid structure.[in] us: A dimensional unit scaling type[in] diag: Diagnostics[in] time: Model timecs: Control structure[out] passive: Copy of passiveModewaves: Wave CS
-
subroutine, public mom_cvmix_kpp::kpp_calculate(CS CS, G G, GV GV, US US, h h, uStar uStar, buoyFlux buoyFlux, Kt Kt, Ks Ks, Kv Kv, nonLocalTransHeat nonLocalTransHeat, nonLocalTransScalar nonLocalTransScalar, waves waves) KPP vertical diffusivity/viscosity and non-local tracer transport.
- Parameters
cs: Control structure[in] g: Ocean grid[in] gv: Ocean vertical grid[in] us: A dimensional unit scaling typewaves: Wave CS[in] h: Layer/level thicknesses [H ~> m or kg m-2][in] ustar: Surface friction velocity [Z T-1 ~> m s-1][in] buoyflux: Surface buoyancy flux [L2 T-3 ~> m2 s-3][inout] kt: (in) Vertical diffusivity of heat w/o KPP (out) Vertical diffusivity including KPP [Z2 T-1 ~> m2 s-1][inout] ks: (in) Vertical diffusivity of salt w/o KPP (out) Vertical diffusivity including KPP [Z2 T-1 ~> m2 s-1][inout] kv: (in) Vertical viscosity w/o KPP (out) Vertical viscosity including KPP [Z2 T-1 ~> m2 s-1][inout] nonlocaltransheat: Temp non-local transport [m s-1][inout] nonlocaltransscalar: scalar non-local transport [m s-1]
-
subroutine, public mom_cvmix_kpp::kpp_compute_bld(CS CS, G G, GV GV, US US, h h, Temp Temp, Salt Salt, u u, v v, EOS EOS, uStar uStar, buoyFlux buoyFlux, Waves Waves) Compute OBL depth.
- Parameters
cs: Control structure[inout] g: Ocean grid[in] gv: Ocean vertical grid[in] us: A dimensional unit scaling type[in] h: Layer/level thicknesses [H ~> m or kg m-2][in] temp: potential/cons temp [degC][in] salt: Salinity [ppt][in] u: Velocity i-component [m s-1][in] v: Velocity j-component [m s-1]eos: Equation of state[in] ustar: Surface friction velocity [Z T-1 ~> m s-1][in] buoyflux: Surface buoyancy flux [L2 T-3 ~> m2 s-3]waves: Wave CS
-
subroutine
kpp_smooth_bld(CS CS, G G, GV GV, h h)¶ Apply a 1-1-4-1-1 Laplacian filter one time on BLD to reduce any horizontal two-grid-point noise.
- Parameters
cs: Control structure[inout] g: Ocean grid[in] gv: Ocean vertical grid[in] h: Layer/level thicknesses [H ~> m or kg m-2]
-
subroutine, public mom_cvmix_kpp::kpp_get_bld(CS CS, BLD BLD, G G) Copies KPP surface boundary layer depth into BLD.
- Parameters
cs: Control structure for this module[in] g: Grid structure[inout] bld: bnd. layer depth [m]
-
subroutine, public mom_cvmix_kpp::kpp_nonlocaltransport_temp(CS CS, G G, GV GV, h h, nonLocalTrans nonLocalTrans, surfFlux surfFlux, dt dt, scalar scalar, C_p C_p) Apply KPP non-local transport of surface fluxes for temperature.
- Parameters
[in] cs: Control structure[in] g: Ocean grid[in] gv: Ocean vertical grid[in] h: Layer/level thickness [H ~> m or kg m-2][in] nonlocaltrans: Non-local transport [nondim][in] surfflux: Surface flux of scalar [conc H s-1 ~> conc m s-1 or conc kg m-2 s-1][in] dt: Time-step [s][inout] scalar: temperature[in] c_p: Seawater specific heat capacity [J kg-1 degC-1]
-
subroutine, public mom_cvmix_kpp::kpp_nonlocaltransport_saln(CS CS, G G, GV GV, h h, nonLocalTrans nonLocalTrans, surfFlux surfFlux, dt dt, scalar scalar) Apply KPP non-local transport of surface fluxes for salinity. This routine is a useful prototype for other material tracers.
- Parameters
[in] cs: Control structure[in] g: Ocean grid[in] gv: Ocean vertical grid[in] h: Layer/level thickness [H ~> m or kg m-2][in] nonlocaltrans: Non-local transport [nondim][in] surfflux: Surface flux of scalar [conc H s-1 ~> conc m s-1 or conc kg m-2 s-1][in] dt: Time-step [s][inout] scalar: Scalar (scalar units [conc])
-
subroutine, public mom_cvmix_kpp::kpp_end(CS CS) Clear pointers, deallocate memory.
- Parameters
cs: Control structure
Variables
-
integer, parameter, private mom_cvmix_kpp::nlt_shape_cvmix = 0 Use the CVMix profile.
-
integer, parameter, private mom_cvmix_kpp::nlt_shape_linear = 1 Linear, \( G(\sigma) = 1-\sigma \).
-
integer, parameter, private mom_cvmix_kpp::nlt_shape_parabolic = 2 Parabolic, \( G(\sigma) = (1-\sigma)^2 \).
-
integer, parameter, private mom_cvmix_kpp::nlt_shape_cubic = 3 Cubic, \( G(\sigma) = 1 + (2\sigma-3) \sigma^2\).
-
integer, parameter, private mom_cvmix_kpp::nlt_shape_cubic_lmd = 4 Original shape, \( G(\sigma) = \frac{27}{4} \sigma (1-\sigma)^2 \).
-
integer, parameter, private mom_cvmix_kpp::sw_method_all_sw = 0 Use all shortwave radiation.
-
integer, parameter, private mom_cvmix_kpp::sw_method_mxl_sw = 1 Use shortwave radiation absorbed in mixing layer.
-
integer, parameter, private mom_cvmix_kpp::sw_method_lv1_sw = 2 Use shortwave radiation absorbed in layer 1.
-
integer, parameter, private mom_cvmix_kpp::lt_k_constant = 1 Constant enhance K through column.
-
integer, parameter, private mom_cvmix_kpp::lt_k_scaled = 2 Enhance K scales with G(sigma)
-
integer, parameter, private mom_cvmix_kpp::lt_k_mode_constant = 1 Prescribed enhancement for K.
-
integer, parameter, private mom_cvmix_kpp::lt_k_mode_vr12 = 2 Enhancement for K based on.
-
integer, parameter, private mom_cvmix_kpp::lt_k_mode_rw16 = 3 Enhancement for K based on.
-
integer, parameter, private mom_cvmix_kpp::lt_vt2_mode_constant = 1 Prescribed enhancement for Vt2.
-
integer, parameter, private mom_cvmix_kpp::lt_vt2_mode_vr12 = 2 Enhancement for Vt2 based on.
-
integer, parameter, private mom_cvmix_kpp::lt_vt2_mode_rw16 = 3 Enhancement for Vt2 based on.
-
integer, parameter, private mom_cvmix_kpp::lt_vt2_mode_lf17 = 4 Enhancement for Vt2 based on.
-
-
namespace
mom_cvmix_shear¶ Interface to CVMix interior shear schemes.
Unnamed Group
-
character(len=40) mom_cvmix_shear::mdl = "MOM_CVMix_shear" This module’s name.
-
subroutine, public mom_cvmix_shear::calculate_cvmix_shear(u_H u_H, v_H v_H, h h, tv tv, kd kd, kv kv, G G, GV GV, US US, CS CS) Subroutine for calculating (internal) vertical diffusivities/viscosities.
- Parameters
[in] g: Grid structure.[in] gv: Vertical grid structure.[in] us: A dimensional unit scaling type[in] u_h: Initial zonal velocity on T points [m s-1].[in] v_h: Initial meridional velocity on T points [m s-1].[in] h: Layer thickness [H ~> m or kg m-2].[in] tv: Thermodynamics structure.[out] kd: The vertical diffusivity at each interface (not layer!) [Z2 T-1 ~> m2 s-1].[out] kv: The vertical viscosity at each interface (not layer!) [Z2 T-1 ~> m2 s-1].cs: The control structure returned by a previous call to CVMix_shear_init.
-
logical function, public mom_cvmix_shear::cvmix_shear_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS) Initialized the CVMix internal shear mixing routine.
- Note
*This is where we test to make sure multiple internal shear mixing routines (including JHL) are not enabled at the same time. (returns) CVMix_shear_init - True if module is to be used, False otherwise
- Parameters
[in] time: The current time.[in] g: Grid structure.[in] gv: Vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: Run-time parameter file handle[inout] diag: Diagnostics control structure.cs: This module’s control structure.
-
logical function, public mom_cvmix_shear::cvmix_shear_is_used(param_file param_file) Reads the parameters “LMD94” and “PP81” and returns state. This function allows other modules to know whether this parameterization will be used without needing to duplicate the log entry.
- Parameters
[in] param_file: Run-time parameter files handle.
-
subroutine, public mom_cvmix_shear::cvmix_shear_end(CS CS) Clear pointers and dealocate memory.
- Parameters
cs: Control structure for this module that will be deallocated in this subroutine
-
-
namespace
mom_debugging¶ Provides checksumming functions for debugging.
This module contains subroutines that perform various error checking and debugging functions for MOM6. This routine is similar to it counterpart in the SIS2 code, except for the use of the ocean_grid_type and by keeping them separate we retain the ability to set up MOM6 and SIS2 debugging separately.
Functions
-
subroutine, public mom_debugging::mom_debugging_init(param_file param_file) MOM_debugging_init initializes the MOM_debugging module, and sets the parameterts that control which checks are active for MOM6.
- Parameters
[in] param_file: A structure to parse for run-time parameters
-
subroutine
check_redundant_vc3d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, is is, ie ie, js js, je je, direction direction)¶ Check for consistency between the duplicated points of a 3-D C-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector to be checked for consistency[in] v_comp: The u-component of the vector to be checked for consistency[in] is: The starting i-index to check[in] ie: The ending i-index to check[in] js: The starting j-index to check[in] je: The ending j-index to check[in] direction: the direction flag to be passed to pass_vector
-
subroutine
check_redundant_vc2d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, is is, ie ie, js js, je je, direction direction)¶ Check for consistency between the duplicated points of a 2-D C-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector to be checked for consistency[in] v_comp: The u-component of the vector to be checked for consistency[in] is: The starting i-index to check[in] ie: The ending i-index to check[in] js: The starting j-index to check[in] je: The ending j-index to check[in] direction: the direction flag to be passed to pass_vector
-
subroutine
check_redundant_sb3d(mesg mesg, array array, G G, is is, ie ie, js js, je je)¶ Check for consistency between the duplicated points of a 3-D scalar at corner points.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] array: The array to be checked for consistency[in] is: The starting i-index to check[in] ie: The ending i-index to check[in] js: The starting j-index to check[in] je: The ending j-index to check
-
subroutine
check_redundant_sb2d(mesg mesg, array array, G G, is is, ie ie, js js, je je)¶ Check for consistency between the duplicated points of a 2-D scalar at corner points.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] array: The array to be checked for consistency[in] is: The starting i-index to check[in] ie: The ending i-index to check[in] js: The starting j-index to check[in] je: The ending j-index to check
-
subroutine
check_redundant_vb3d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, is is, ie ie, js js, je je, direction direction)¶ Check for consistency between the duplicated points of a 3-D B-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector to be checked for consistency[in] v_comp: The v-component of the vector to be checked for consistency[in] is: The starting i-index to check[in] ie: The ending i-index to check[in] js: The starting j-index to check[in] je: The ending j-index to check[in] direction: the direction flag to be passed to pass_vector
-
subroutine
check_redundant_vb2d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, is is, ie ie, js js, je je, direction direction)¶ Check for consistency between the duplicated points of a 2-D B-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector to be checked for consistency[in] v_comp: The v-component of the vector to be checked for consistency[in] is: The starting i-index to check[in] ie: The ending i-index to check[in] js: The starting j-index to check[in] je: The ending j-index to check[in] direction: the direction flag to be passed to pass_vector
-
subroutine
check_redundant_st3d(mesg mesg, array array, G G, is is, ie ie, js js, je je)¶ Check for consistency between the duplicated points of a 3-D scalar at tracer points.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] array: The array to be checked for consistency[in] is: The starting i-index to check[in] ie: The ending i-index to check[in] js: The starting j-index to check[in] je: The ending j-index to check
-
subroutine
check_redundant_st2d(mesg mesg, array array, G G, is is, ie ie, js js, je je)¶ Check for consistency between the duplicated points of a 2-D scalar at tracer points.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] array: The array to be checked for consistency[in] is: The starting i-index to check[in] ie: The ending i-index to check[in] js: The starting j-index to check[in] je: The ending j-index to check
-
subroutine
check_redundant_vt3d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, is is, ie ie, js js, je je, direction direction)¶ Check for consistency between the duplicated points of a 3-D A-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector to be checked for consistency[in] v_comp: The v-component of the vector to be checked for consistency[in] is: The starting i-index to check[in] ie: The ending i-index to check[in] js: The starting j-index to check[in] je: The ending j-index to check[in] direction: the direction flag to be passed to pass_vector
-
subroutine
check_redundant_vt2d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, is is, ie ie, js js, je je, direction direction)¶ Check for consistency between the duplicated points of a 2-D A-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector to be checked for consistency[in] v_comp: The v-component of the vector to be checked for consistency[in] is: The starting i-index to check[in] ie: The ending i-index to check[in] js: The starting j-index to check[in] je: The ending j-index to check[in] direction: the direction flag to be passed to pass_vector
-
subroutine
chksum_vec_c3d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, halos halos, scalars scalars)¶ Do a checksum and redundant point check on a 3d C-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector[in] v_comp: The v-component of the vector[in] halos: The width of halos to check (default 0)[in] scalars: If true this is a pair of scalars that are being checked.
-
subroutine
chksum_vec_c2d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, halos halos, scalars scalars)¶ Do a checksum and redundant point check on a 2d C-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector[in] v_comp: The v-component of the vector[in] halos: The width of halos to check (default 0)[in] scalars: If true this is a pair of scalars that are being checked.
-
subroutine
chksum_vec_b3d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, halos halos, scalars scalars)¶ Do a checksum and redundant point check on a 3d B-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector[in] v_comp: The v-component of the vector[in] halos: The width of halos to check (default 0)[in] scalars: If true this is a pair of scalars that are being checked.
-
subroutine
chksum_vec_b2d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, halos halos, scalars scalars, symmetric symmetric)¶ - Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector[in] v_comp: The v-component of the vector[in] halos: The width of halos to check (default 0)[in] scalars: If true this is a pair of scalars that are being checked.[in] symmetric: If true, do the checksums on the full symmetric computational domain.
-
subroutine
chksum_vec_a3d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, halos halos, scalars scalars)¶ Do a checksum and redundant point check on a 3d C-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector[in] v_comp: The v-component of the vector[in] halos: The width of halos to check (default 0)[in] scalars: If true this is a pair of scalars that are being checked.
-
subroutine
chksum_vec_a2d(mesg mesg, u_comp u_comp, v_comp v_comp, G G, halos halos, scalars scalars)¶ Do a checksum and redundant point check on a 2d C-grid vector.
- Parameters
[in] mesg: An identifying message[inout] g: The ocean’s grid structure[in] u_comp: The u-component of the vector[in] v_comp: The v-component of the vector[in] halos: The width of halos to check (default 0)[in] scalars: If true this is a pair of scalars that are being checked.
-
real function, public mom_debugging::totalstuff(HI HI, hThick hThick, areaT areaT, stuff stuff) This function returns the sum over computational domain of all processors of hThick*stuff, where stuff is a 3-d array at tracer points.
- Return
the globally integrated amoutn of stuff
- Parameters
[in] hi: A horizontal index type[in] hthick: The array of thicknesses to use as weights[in] areat: The array of cell areas [m2][in] stuff: The array of stuff to be summed
-
subroutine, public mom_debugging::totaltands(HI HI, hThick hThick, areaT areaT, temperature temperature, salinity salinity, mesg mesg) This subroutine display the total thickness, temperature and salinity as well as the change since the last call.
- Parameters
[in] hi: A horizontal index type[in] hthick: The array of thicknesses to use as weights[in] areat: The array of cell areas [m2][in] temperature: The temperature field to sum[in] salinity: The salinity field to sum[in] mesg: An identifying message
-
logical function, public mom_debugging::check_column_integral(nk nk, field field, known_answer known_answer) Returns false if the column integral of a given quantity is within roundoff.
- Parameters
[in] nk: Number of levels in column[in] field: Field to be summed[in] known_answer: If present is the expected sum, If missing, assumed zero
-
logical function, public mom_debugging::check_column_integrals(nk_1 nk_1, field_1 field_1, nk_2 nk_2, field_2 field_2, missing_value missing_value) Returns false if the column integrals of two given quantities are within roundoff of each other.
- Parameters
[in] nk_1: Number of levels in field 1[in] nk_2: Number of levels in field 2[in] field_1: First field to be summed[in] field_2: Second field to be summed[in] missing_value: If column contains missing values, mask them from the sum
Variables
-
integer
max_redundant_prints= 100¶ Maximum number of times to write redundant messages.
-
integer, dimension(3) mom_debugging::redundant_prints = 0 Counters for controlling redundant printing.
-
logical
debug= .false.¶ Write out verbose debugging data.
-
logical
debug_chksums= .true.¶ Perform checksums on arrays.
-
logical
debug_redundant= .true.¶ Check redundant values on PE boundaries.
-
-
namespace
mom_diabatic_aux¶ Provides functions for some diabatic processes such as fraxil, brine rejection, tendency due to surface flux divergence.
This module contains the subroutines that, along with the subroutines that it calls, implements diapycnal mass and momentum fluxes and a bulk mixed layer. The diapycnal diffusion can be used without the bulk mixed layer.
diabatic first determines the (diffusive) diapycnal mass fluxes based on the convergence of the buoyancy fluxes within each layer. The dual-stream entrainment scheme of MacDougall and Dewar (JPO, 1997) is used for combined diapycnal advection and diffusion, calculated implicitly and potentially with the Richardson number dependent mixing, as described by Hallberg (MWR, 2000). Diapycnal advection is fundamentally the residual of diapycnal diffusion, so the fully implicit upwind differencing scheme that is used is entirely appropriate. The downward buoyancy flux in each layer is determined from an implicit calculation based on the previously calculated flux of the layer above and an estimated flux in the layer below. This flux is subject to the following conditions: (1) the flux in the top and bottom layers are set by the boundary conditions, and (2) no layer may be driven below an Angstrom thick- ness. If there is a bulk mixed layer, the buffer layer is treat- ed as a fixed density layer with vanishingly small diffusivity.
diabatic takes 5 arguments: the two velocities (u and v), the thicknesses (h), a structure containing the forcing fields, and the length of time over which to act (dt). The velocities and thickness are taken as inputs and modified within the subroutine. There is no limit on the time step.
Unnamed Group
-
integer
id_clock_uv_at_h¶ CPU time clock IDs.
-
integer
id_clock_frazil¶ CPU time clock IDs.
-
subroutine, public mom_diabatic_aux::make_frazil(h h, tv tv, G G, GV GV, CS CS, p_surf p_surf, halo halo) Frazil formation keeps the temperature above the freezing point. This subroutine warms any water that is colder than the (currently surface) freezing point up to the freezing point and accumulates the required heat (in J m-2) in tvfrazil.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][inout] tv: Structure containing pointers to any available thermodynamic fields.[in] cs: The control structure returned by a previous call to diabatic_aux_init.[in] p_surf: The pressure at the ocean surface [Pa].[in] halo: Halo width over which to calculate frazil
-
subroutine, public mom_diabatic_aux::differential_diffuse_t_s(h h, tv tv, visc visc, dt dt, G G, GV GV) This subroutine applies double diffusion to T & S, assuming no diapycal mass fluxes, using a simple triadiagonal solver.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][inout] tv: Structure containing pointers to any available thermodynamic fields.[in] visc: Structure containing vertical viscosities, bottom boundary layer properies, and related fields.[in] dt: Time increment [T ~> s].
-
subroutine, public mom_diabatic_aux::adjust_salt(h h, tv tv, G G, GV GV, CS CS, halo halo) This subroutine keeps salinity from falling below a small but positive threshold. This usually occurs when the ice model attempts to extract more salt then is actually available to it from the ocean.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][inout] tv: Structure containing pointers to any available thermodynamic fields.[in] cs: The control structure returned by a previous call to diabatic_aux_init.[in] halo: Halo width over which to work
-
subroutine, public mom_diabatic_aux::insert_brine(h h, tv tv, G G, GV GV, fluxes fluxes, nkmb nkmb, CS CS, dt dt, id_brine_lay id_brine_lay) Insert salt from brine rejection into the first layer below the mixed layer which both contains mass and in which the change in layer density remains stable after the addition of salt via brine rejection.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][inout] tv: Structure containing pointers to any available thermodynamic fields[in] fluxes: A structure of thermodynamic surface fluxes[in] nkmb: The number of layers in the mixed and buffer layers[in] cs: The control structure returned by a previous call to diabatic_aux_init[in] dt: The thermodynamic time step [s].[in] id_brine_lay: The handle for a diagnostic of which layer receivees the brine.
-
subroutine, public mom_diabatic_aux::tridiagts(G G, GV GV, is is, ie ie, js js, je je, hold hold, ea ea, eb eb, T T, S S) This is a simple tri-diagonal solver for T and S. “Simple” means it only uses arrays hold, ea and eb.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] is: The start i-index to work on.[in] ie: The end i-index to work on.[in] js: The start j-index to work on.[in] je: The end j-index to work on.[in] hold: The layer thicknesses before entrainment, [H ~> m or kg m-2].[in] ea: The amount of fluid entrained from the layer above within this time step [H ~> m or kg m-2].[in] eb: The amount of fluid entrained from the layer below within this time step [H ~> m or kg m-2].[inout] t: Layer potential temperatures [degC].[inout] s: Layer salinities [ppt].
-
subroutine, public mom_diabatic_aux::find_uv_at_h(u u, v v, h h, u_h u_h, v_h v_h, G G, GV GV, ea ea, eb eb) This subroutine calculates u_h and v_h (velocities at thickness points), optionally using the entrainment amounts passed in as arguments.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] u: The zonal velocity [m s-1][in] v: The meridional velocity [m s-1][in] h: Layer thicknesses [H ~> m or kg m-2][out] u_h: Zonal velocity interpolated to h points [m s-1].[out] v_h: Meridional velocity interpolated to h points [m s-1].[in] ea: The amount of fluid entrained from the layer[in] eb: The amount of fluid entrained from the layer
-
subroutine, public mom_diabatic_aux::set_pen_shortwave(optics optics, fluxes fluxes, G G, GV GV, CS CS, opacity_CSp opacity_CSp, tracer_flow_CSp tracer_flow_CSp) - Parameters
optics: An optics structure that has will contain information about shortwave fluxes and absorption.[inout] fluxes: points to forcing fields unused fields have NULL ptrs[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.cs: Control structure for diabatic_auxopacity_csp: The control structure for the opacity module.tracer_flow_csp: A pointer to the control structure of the tracer modules.
-
subroutine, public mom_diabatic_aux::diagnosemldbydensitydifference(id_MLD id_MLD, h h, tv tv, densityDiff densityDiff, G G, GV GV, US US, diagPtr diagPtr, id_N2subML id_N2subML, id_MLDsq id_MLDsq, dz_subML dz_subML) Diagnose a mixed layer depth (MLD) determined by a given density difference with the surface. This routine is appropriate in MOM_diabatic_driver due to its position within the time stepping.
- Parameters
[in] g: Grid type[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[in] id_mld: Handle (ID) of MLD diagnostic[in] h: Layer thickness [H ~> m or kg m-2][in] tv: Structure containing pointers to any available thermodynamic fields.[in] densitydiff: Density difference to determine MLD [kg m-3]diagptr: Diagnostics structure[in] id_n2subml: Optional handle (ID) of subML stratification[in] id_mldsq: Optional handle (ID) of squared MLD[in] dz_subml: The distance over which to calculate N2subML or 50 m if missing [Z ~> m]
-
subroutine, public mom_diabatic_aux::applyboundaryfluxesinout(CS CS, G G, GV GV, US US, dt dt, fluxes fluxes, optics optics, nsw nsw, h h, tv tv, aggregate_FW_forcing aggregate_FW_forcing, evap_CFL_limit evap_CFL_limit, minimum_forcing_depth minimum_forcing_depth, cTKE cTKE, dSV_dT dSV_dT, dSV_dS dSV_dS, SkinBuoyFlux SkinBuoyFlux) Update the thickness, temperature, and salinity due to thermodynamic boundary forcing (contained in fluxes type) applied to h, tvT and tvS, and calculate the TKE implications of this heating.
- Parameters
cs: Control structure for diabatic_aux[in] g: Grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[in] dt: Time-step over which forcing is applied [s][inout] fluxes: Surface fluxes containeroptics: Optical properties container[in] nsw: The number of frequency bands of penetrating shortwave radiation[inout] h: Layer thickness [H ~> m or kg m-2][inout] tv: Structure containing pointers to any available thermodynamic fields.[in] aggregate_fw_forcing: If False, treat in/out fluxes separately.[in] evap_cfl_limit: The largest fraction of a layer that can be evaporated in one time-step [nondim].[in] minimum_forcing_depth: The smallest depth over which heat and freshwater fluxes is applied [m].[out] ctke: Turbulent kinetic energy requirement to mix[out] dsv_dt: Partial derivative of specific volume with[out] dsv_ds: Partial derivative of specific volume with[out] skinbuoyflux: Buoyancy flux at surface [Z2 T-3 ~> m2 s-3].
-
subroutine, public mom_diabatic_aux::diabatic_aux_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS, useALEalgorithm useALEalgorithm, use_ePBL use_ePBL) This subroutine initializes the parameters and control structure of the diabatic_aux module.
- Parameters
[in] time: The current model time.[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters[inout] diag: A structure used to regulate diagnostic outputcs: A pointer to the control structure for the diabatic_aux module, which is initialized here.[in] usealealgorithm: If true, use the ALE algorithm rather than layered mode.[in] use_epbl: If true, use the implicit energetics planetary boundary layer scheme to determine the diffusivity in the surface boundary layer.
-
subroutine, public mom_diabatic_aux::diabatic_aux_end(CS CS) This subroutine initializes the control structure and any related memory for the diabatic_aux module.
- Parameters
cs: The control structure returned by a previous call to diabatic_aux_init; it is deallocated here.
-
integer
-
namespace
mom_diabatic_driver¶ This routine drives the diabatic/dianeutral physics for MOM.
By Robert Hallberg, Alistair Adcroft, and Stephen Griffies
This program contains the subroutine that, along with the subroutines that it calls, implements diapycnal mass and momentum fluxes and a bulk mixed layer. The diapycnal diffusion can be used without the bulk mixed layer.
Unnamed Group
-
integer
id_clock_entrain¶ This subroutine imposes the diapycnal mass fluxes and the accompanying diapycnal advection of momentum and tracers.
- Parameters
[inout] g: ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] u: zonal velocity [m s-1][inout] v: meridional velocity [m s-1][inout] h: thickness [H ~> m or kg m-2][inout] tv: points to thermodynamic fields unused have NULL ptrshml: Active mixed layer depth [m][inout] fluxes: points to forcing fields unused fields have NULL ptrs[inout] visc: vertical viscosities, BBL properies, and[inout] adp: related points to accelerations in momentum equations, to enable the later derived diagnostics, like energy budgets[inout] cdp: points to terms in continuity equations[in] dt: time increment [s][in] time_end: Time at the end of the intervalcs: module control structurewaves: Surface gravity waves
-
integer
id_clock_mixedlayer¶ This subroutine imposes the diapycnal mass fluxes and the accompanying diapycnal advection of momentum and tracers.
- Parameters
[inout] g: ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] u: zonal velocity [m s-1][inout] v: meridional velocity [m s-1][inout] h: thickness [H ~> m or kg m-2][inout] tv: points to thermodynamic fields unused have NULL ptrshml: Active mixed layer depth [m][inout] fluxes: points to forcing fields unused fields have NULL ptrs[inout] visc: vertical viscosities, BBL properies, and[inout] adp: related points to accelerations in momentum equations, to enable the later derived diagnostics, like energy budgets[inout] cdp: points to terms in continuity equations[in] dt: time increment [s][in] time_end: Time at the end of the intervalcs: module control structurewaves: Surface gravity waves
-
integer
id_clock_set_diffusivity¶ This subroutine imposes the diapycnal mass fluxes and the accompanying diapycnal advection of momentum and tracers.
- Parameters
[inout] g: ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] u: zonal velocity [m s-1][inout] v: meridional velocity [m s-1][inout] h: thickness [H ~> m or kg m-2][inout] tv: points to thermodynamic fields unused have NULL ptrshml: Active mixed layer depth [m][inout] fluxes: points to forcing fields unused fields have NULL ptrs[inout] visc: vertical viscosities, BBL properies, and[inout] adp: related points to accelerations in momentum equations, to enable the later derived diagnostics, like energy budgets[inout] cdp: points to terms in continuity equations[in] dt: time increment [s][in] time_end: Time at the end of the intervalcs: module control structurewaves: Surface gravity waves
-
integer
id_clock_tracers¶ This subroutine imposes the diapycnal mass fluxes and the accompanying diapycnal advection of momentum and tracers.
- Parameters
[inout] g: ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] u: zonal velocity [m s-1][inout] v: meridional velocity [m s-1][inout] h: thickness [H ~> m or kg m-2][inout] tv: points to thermodynamic fields unused have NULL ptrshml: Active mixed layer depth [m][inout] fluxes: points to forcing fields unused fields have NULL ptrs[inout] visc: vertical viscosities, BBL properies, and[inout] adp: related points to accelerations in momentum equations, to enable the later derived diagnostics, like energy budgets[inout] cdp: points to terms in continuity equations[in] dt: time increment [s][in] time_end: Time at the end of the intervalcs: module control structurewaves: Surface gravity waves
-
integer
id_clock_tridiag¶ This subroutine imposes the diapycnal mass fluxes and the accompanying diapycnal advection of momentum and tracers.
- Parameters
[inout] g: ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] u: zonal velocity [m s-1][inout] v: meridional velocity [m s-1][inout] h: thickness [H ~> m or kg m-2][inout] tv: points to thermodynamic fields unused have NULL ptrshml: Active mixed layer depth [m][inout] fluxes: points to forcing fields unused fields have NULL ptrs[inout] visc: vertical viscosities, BBL properies, and[inout] adp: related points to accelerations in momentum equations, to enable the later derived diagnostics, like energy budgets[inout] cdp: points to terms in continuity equations[in] dt: time increment [s][in] time_end: Time at the end of the intervalcs: module control structurewaves: Surface gravity waves
-
integer
id_clock_pass¶ This subroutine imposes the diapycnal mass fluxes and the accompanying diapycnal advection of momentum and tracers.
- Parameters
[inout] g: ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] u: zonal velocity [m s-1][inout] v: meridional velocity [m s-1][inout] h: thickness [H ~> m or kg m-2][inout] tv: points to thermodynamic fields unused have NULL ptrshml: Active mixed layer depth [m][inout] fluxes: points to forcing fields unused fields have NULL ptrs[inout] visc: vertical viscosities, BBL properies, and[inout] adp: related points to accelerations in momentum equations, to enable the later derived diagnostics, like energy budgets[inout] cdp: points to terms in continuity equations[in] dt: time increment [s][in] time_end: Time at the end of the intervalcs: module control structurewaves: Surface gravity waves
-
integer
id_clock_sponge¶ This subroutine imposes the diapycnal mass fluxes and the accompanying diapycnal advection of momentum and tracers.
- Parameters
[inout] g: ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] u: zonal velocity [m s-1][inout] v: meridional velocity [m s-1][inout] h: thickness [H ~> m or kg m-2][inout] tv: points to thermodynamic fields unused have NULL ptrshml: Active mixed layer depth [m][inout] fluxes: points to forcing fields unused fields have NULL ptrs[inout] visc: vertical viscosities, BBL properies, and[inout] adp: related points to accelerations in momentum equations, to enable the later derived diagnostics, like energy budgets[inout] cdp: points to terms in continuity equations[in] dt: time increment [s][in] time_end: Time at the end of the intervalcs: module control structurewaves: Surface gravity waves
-
integer
id_clock_geothermal¶ This subroutine imposes the diapycnal mass fluxes and the accompanying diapycnal advection of momentum and tracers.
- Parameters
[inout] g: ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] u: zonal velocity [m s-1][inout] v: meridional velocity [m s-1][inout] h: thickness [H ~> m or kg m-2][inout] tv: points to thermodynamic fields unused have NULL ptrshml: Active mixed layer depth [m][inout] fluxes: points to forcing fields unused fields have NULL ptrs[inout] visc: vertical viscosities, BBL properies, and[inout] adp: related points to accelerations in momentum equations, to enable the later derived diagnostics, like energy budgets[inout] cdp: points to terms in continuity equations[in] dt: time increment [s][in] time_end: Time at the end of the intervalcs: module control structurewaves: Surface gravity waves
-
integer
id_clock_differential_diff¶ This subroutine imposes the diapycnal mass fluxes and the accompanying diapycnal advection of momentum and tracers.
- Parameters
[inout] g: ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] u: zonal velocity [m s-1][inout] v: meridional velocity [m s-1][inout] h: thickness [H ~> m or kg m-2][inout] tv: points to thermodynamic fields unused have NULL ptrshml: Active mixed layer depth [m][inout] fluxes: points to forcing fields unused fields have NULL ptrs[inout] visc: vertical viscosities, BBL properies, and[inout] adp: related points to accelerations in momentum equations, to enable the later derived diagnostics, like energy budgets[inout] cdp: points to terms in continuity equations[in] dt: time increment [s][in] time_end: Time at the end of the intervalcs: module control structurewaves: Surface gravity waves
-
integer
id_clock_remap¶ This subroutine imposes the diapycnal mass fluxes and the accompanying diapycnal advection of momentum and tracers.
- Parameters
[inout] g: ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] u: zonal velocity [m s-1][inout] v: meridional velocity [m s-1][inout] h: thickness [H ~> m or kg m-2][inout] tv: points to thermodynamic fields unused have NULL ptrshml: Active mixed layer depth [m][inout] fluxes: points to forcing fields unused fields have NULL ptrs[inout] visc: vertical viscosities, BBL properies, and[inout] adp: related points to accelerations in momentum equations, to enable the later derived diagnostics, like energy budgets[inout] cdp: points to terms in continuity equations[in] dt: time increment [s][in] time_end: Time at the end of the intervalcs: module control structurewaves: Surface gravity waves
-
integer
id_clock_kpp¶ This subroutine imposes the diapycnal mass fluxes and the accompanying diapycnal advection of momentum and tracers.
- Parameters
[inout] g: ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] u: zonal velocity [m s-1][inout] v: meridional velocity [m s-1][inout] h: thickness [H ~> m or kg m-2][inout] tv: points to thermodynamic fields unused have NULL ptrshml: Active mixed layer depth [m][inout] fluxes: points to forcing fields unused fields have NULL ptrs[inout] visc: vertical viscosities, BBL properies, and[inout] adp: related points to accelerations in momentum equations, to enable the later derived diagnostics, like energy budgets[inout] cdp: points to terms in continuity equations[in] dt: time increment [s][in] time_end: Time at the end of the intervalcs: module control structurewaves: Surface gravity waves
-
subroutine, public mom_diabatic_driver::diabatic(u u, v v, h h, tv tv, Hml Hml, fluxes fluxes, visc visc, ADp ADp, CDp CDp, dt dt, Time_end Time_end, G G, GV GV, US US, CS CS, WAVES WAVES) This subroutine imposes the diapycnal mass fluxes and the accompanying diapycnal advection of momentum and tracers.
- Parameters
[inout] g: ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] u: zonal velocity [m s-1][inout] v: meridional velocity [m s-1][inout] h: thickness [H ~> m or kg m-2][inout] tv: points to thermodynamic fields unused have NULL ptrshml: Active mixed layer depth [m][inout] fluxes: points to forcing fields unused fields have NULL ptrs[inout] visc: vertical viscosities, BBL properies, and[inout] adp: related points to accelerations in momentum equations, to enable the later derived diagnostics, like energy budgets[inout] cdp: points to terms in continuity equations[in] dt: time increment [s][in] time_end: Time at the end of the intervalcs: module control structurewaves: Surface gravity waves
-
subroutine
diabatic_ale_legacy(u u, v v, h h, tv tv, Hml Hml, fluxes fluxes, visc visc, ADp ADp, CDp CDp, dt dt, Time_end Time_end, G G, GV GV, US US, CS CS, WAVES WAVES)¶ Applies diabatic forcing and diapycnal mixing of temperature, salinity and other tracers for use with an ALE algorithm. This version uses an older set of algorithms compared with diabatic_ALE.
- Parameters
[inout] g: ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] u: zonal velocity [m s-1][inout] v: meridional velocity [m s-1][inout] h: thickness [H ~> m or kg m-2][inout] tv: points to thermodynamic fields unused have NULL ptrshml: Active mixed layer depth [m][inout] fluxes: points to forcing fields unused fields have NULL ptrs[inout] visc: vertical viscosities, BBL properies, and[inout] adp: related points to accelerations in momentum equations, to enable the later derived diagnostics, like energy budgets[inout] cdp: points to terms in continuity equations[in] dt: time increment [s][in] time_end: Time at the end of the intervalcs: module control structurewaves: Surface gravity waves
-
subroutine
diabatic_ale(u u, v v, h h, tv tv, Hml Hml, fluxes fluxes, visc visc, ADp ADp, CDp CDp, dt dt, Time_end Time_end, G G, GV GV, US US, CS CS, Waves Waves)¶ This subroutine imposes the diapycnal mass fluxes and the accompanying diapycnal advection of momentum and tracers.
- Parameters
[inout] g: ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] u: zonal velocity [m s-1][inout] v: meridional velocity [m s-1][inout] h: thickness [H ~> m or kg m-2][inout] tv: points to thermodynamic fields unused have NULL ptrshml: Active mixed layer depth [m][inout] fluxes: points to forcing fields unused fields have NULL ptrs[inout] visc: vertical viscosities, BBL properies, and[inout] adp: related points to accelerations in momentum equations, to enable the later derived diagnostics, like energy budgets[inout] cdp: points to terms in continuity equations[in] dt: time increment [s][in] time_end: Time at the end of the intervalcs: module control structurewaves: Surface gravity waves
-
subroutine
layered_diabatic(u u, v v, h h, tv tv, Hml Hml, fluxes fluxes, visc visc, ADp ADp, CDp CDp, dt dt, Time_end Time_end, G G, GV GV, US US, CS CS, WAVES WAVES)¶ Imposes the diapycnal mass fluxes and the accompanying diapycnal advection of momentum and tracers using the original MOM6 algorithms.
- Parameters
[inout] g: ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] u: zonal velocity [m s-1][inout] v: meridional velocity [m s-1][inout] h: thickness [H ~> m or kg m-2][inout] tv: points to thermodynamic fields unused have NULL ptrshml: Active mixed layer depth [m][inout] fluxes: points to forcing fields unused fields have NULL ptrs[inout] visc: vertical viscosities, BBL properies, and[inout] adp: related points to accelerations in momentum equations, to enable the later derived diagnostics, like energy budgets[inout] cdp: points to terms in continuity equations[in] dt: time increment [s][in] time_end: Time at the end of the intervalcs: module control structurewaves: Surface gravity waves
-
subroutine, public mom_diabatic_driver::extract_diabatic_member(CS CS, opacity_CSp opacity_CSp, optics_CSp optics_CSp, evap_CFL_limit evap_CFL_limit, minimum_forcing_depth minimum_forcing_depth, diabatic_aux_CSp diabatic_aux_CSp) Returns pointers or values of members within the diabatic_CS type. For extensibility, each returned argument is an optional argument.
- Parameters
[in] cs: module control structureopacity_csp: A pointer to be set to the opacity control structureoptics_csp: A pointer to be set to the optics control structure[out] evap_cfl_limit: The largest fraction of a layer that can be evaporated in one time-step [nondim].[out] minimum_forcing_depth: The smallest depth over which heat and freshwater fluxes are applied [m].diabatic_aux_csp: A pointer to be set to the diabatic_aux control structure
-
subroutine, public mom_diabatic_driver::adiabatic(h h, tv tv, fluxes fluxes, dt dt, G G, GV GV, CS CS) Routine called for adiabatic physics.
- Parameters
[inout] g: ocean grid structure[inout] h: thickness [H ~> m or kg m-2][inout] tv: points to thermodynamic fields[inout] fluxes: boundary fluxes[in] dt: time step [s][in] gv: ocean vertical grid structurecs: module control structure
-
subroutine
diagnose_diabatic_diff_tendency(tv tv, h h, temp_old temp_old, saln_old saln_old, dt dt, G G, GV GV, CS CS)¶ This routine diagnoses tendencies from application of diabatic diffusion using ALE algorithm. Note that layer thickness is not altered by diabatic diffusion.
- Parameters
[in] g: ocean grid structure[in] gv: ocean vertical grid structure[in] tv: points to updated thermodynamic fields[in] h: thickness [H ~> m or kg m-2][in] temp_old: temperature prior to diabatic physics[in] saln_old: salinity prior to diabatic physics [ppt][in] dt: time step [s]cs: module control structure
-
subroutine
diagnose_boundary_forcing_tendency(tv tv, h h, temp_old temp_old, saln_old saln_old, h_old h_old, dt dt, G G, GV GV, CS CS)¶ This routine diagnoses tendencies from application of boundary fluxes. These impacts are generally 3d, in particular for penetrative shortwave. Other fluxes contribute 3d in cases when the layers vanish or are very thin, in which case we distribute the flux into k > 1 layers.
- Parameters
[in] g: ocean grid structure[in] gv: ocean vertical grid structure[in] tv: points to updated thermodynamic fields[in] h: thickness after boundary flux application [H ~> m or kg m-2][in] temp_old: temperature prior to boundary flux application [degC][in] saln_old: salinity prior to boundary flux application [ppt][in] h_old: thickness prior to boundary flux application [H ~> m or kg m-2][in] dt: time step [s]cs: module control structure
-
subroutine
diagnose_frazil_tendency(tv tv, h h, temp_old temp_old, dt dt, G G, GV GV, CS CS)¶ This routine diagnoses tendencies for temperature and heat from frazil formation. This routine is called twice from within subroutine diabatic; at start and at end of the diabatic processes. The impacts from frazil are generally a function of depth. Hence, when checking heat budget, be sure to remove HFSIFRAZIL from HFDS in k=1.
- Parameters
[in] g: ocean grid structure[in] gv: ocean vertical grid structurecs: module control structure[in] tv: points to updated thermodynamic fields[in] h: thickness [H ~> m or kg m-2][in] temp_old: temperature prior to frazil formation [degC][in] dt: time step [s]
-
subroutine, public mom_diabatic_driver::adiabatic_driver_init(Time Time, G G, param_file param_file, diag diag, CS CS, tracer_flow_CSp tracer_flow_CSp) A simplified version of diabatic_driver_init that will allow tracer column functions to be called without allowing any of the diabatic processes to be used.
- Parameters
[in] time: current model time[in] g: model grid structure[in] param_file: the file to parse for parameter values[inout] diag: regulates diagnostic outputcs: module control structuretracer_flow_csp: pointer to control structure of the tracer flow control module
-
subroutine, public mom_diabatic_driver::diabatic_driver_init(Time Time, G G, GV GV, US US, param_file param_file, useALEalgorithm useALEalgorithm, diag diag, ADp ADp, CDp CDp, CS CS, tracer_flow_CSp tracer_flow_CSp, sponge_CSp sponge_CSp, ALE_sponge_CSp ALE_sponge_CSp) This routine initializes the diabatic driver module.
- Parameters
time: model time[inout] g: model grid structure[in] gv: model vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: file to parse for parameter values[in] usealealgorithm: logical for whether to use ALE remapping[inout] diag: structure to regulate diagnostic output[inout] adp: pointers to accelerations in momentum equations, to enable diagnostics, like energy budgets[inout] cdp: pointers to terms in continuity equationscs: module control structuretracer_flow_csp: pointer to control structure of the tracer flow control modulesponge_csp: pointer to the sponge module control structureale_sponge_csp: pointer to the ALE sponge module control structure
-
subroutine, public mom_diabatic_driver::diabatic_driver_end(CS CS) Routine to close the diabatic driver module.
- Parameters
cs: module control structure
-
integer
-
namespace
mom_diag_manager_wrapper¶ A simple (very thin) wrapper for register_diag_field to avoid a compiler bug with PGI.
This module simply wraps register_diag_field() from FMS’s diag_manager_mod. We used to be able to import register_diag_field and rename it to register_diag_field_fms with a simple “use, only : register_diag_field_fms => register_diag_field” but PGI 16.5 has a bug that refuses to compile this - earlier versions did work.
Functions
-
integer function mom_diag_manager_wrapper::register_diag_field_array_fms(module_name module_name, field_name field_name, axes axes, init_time init_time, long_name long_name, units units, missing_value missing_value, range range, mask_variant mask_variant, standard_name standard_name, verbose verbose, do_not_log do_not_log, err_msg err_msg, interp_method interp_method, tile_count tile_count, area area, volume volume) An integer handle for a diagnostic array returned by register_diag_field()
- Parameters
[in] module_name: Name of this module, usually “ocean_model” or “ice_shelf_model”[in] field_name: Name of the diagnostic field[in] axes: Container w/ up to 3 integer handles that indicates axes for this field[in] init_time: Time at which a field is first available?[in] long_name: Long name of a field.[in] units: Units of a field.[in] standard_name: Standardized name associated with a field[in] missing_value: A value that indicates missing values.[in] range: Valid range of a variable (not used in MOM?)[in] mask_variant: If true a logical mask must be provided with post_data calls (not used in MOM?)[in] verbose: If true, FMS is verbose (not used in MOM?)[in] do_not_log: If true, do not log something (not used in MOM?)[out] err_msg: String into which an error message might be placed (not used in MOM?)[in] interp_method: If ‘none’ indicates the field should not be interpolated as a scalar[in] tile_count: no clue (not used in MOM?)[in] area: The FMS id of cell area[in] volume: The FMS id of cell volume
-
integer function mom_diag_manager_wrapper::register_diag_field_scalar_fms(module_name module_name, field_name field_name, init_time init_time, long_name long_name, units units, missing_value missing_value, range range, mask_variant mask_variant, standard_name standard_name, verbose verbose, do_not_log do_not_log, err_msg err_msg, interp_method interp_method, tile_count tile_count, area area, volume volume) An integer handle for a diagnostic scalar array returned by register_diag_field()
- Parameters
[in] module_name: Name of this module, usually “ocean_model” or “ice_shelf_model”[in] field_name: Name of the diagnostic field[in] init_time: Time at which a field is first available?[in] long_name: Long name of a field.[in] units: Units of a field.[in] standard_name: Standardized name associated with a field[in] missing_value: A value that indicates missing values.[in] range: Valid range of a variable (not used in MOM?)[in] mask_variant: If true a logical mask must be provided with post_data calls (not used in MOM?)[in] verbose: If true, FMS is verbose (not used in MOM?)[in] do_not_log: If true, do not log something (not used in MOM?)[out] err_msg: String into which an error message might be placed (not used in MOM?)[in] interp_method: If ‘none’ indicates the field should not be interpolated as a scalar[in] tile_count: no clue (not used in MOM?)[in] area: The FMS id of cell area (not used for scalars)[in] volume: The FMS id of cell volume (not used for scalars)
-
-
namespace
mom_diag_mediator¶ The subroutines here provide convenient wrappers to the fms diag_manager interfaces with additional diagnostic capabilies.
Unnamed Group
-
integer
id_clock_diag_mediator¶ Sets up diagnostics axes.
- Parameters
[inout] g: Ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: Parameter file structure[inout] diag_cs: Diagnostics control structure[in] set_vertical: If true or missing, set up vertical axes
-
integer
id_clock_diag_remap¶ Sets up diagnostics axes.
- Parameters
[inout] g: Ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: Parameter file structure[inout] diag_cs: Diagnostics control structure[in] set_vertical: If true or missing, set up vertical axes
-
integer
id_clock_diag_grid_updates¶ Sets up diagnostics axes.
- Parameters
[inout] g: Ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: Parameter file structure[inout] diag_cs: Diagnostics control structure[in] set_vertical: If true or missing, set up vertical axes
-
subroutine, public mom_diag_mediator::set_axes_info(G G, GV GV, US US, param_file param_file, diag_cs diag_cs, set_vertical set_vertical) Sets up diagnostics axes.
- Parameters
[inout] g: Ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: Parameter file structure[inout] diag_cs: Diagnostics control structure[in] set_vertical: If true or missing, set up vertical axes
-
subroutine
set_axes_info_dsamp(G G, GV GV, param_file param_file, diag_cs diag_cs, id_zl_native id_zl_native, id_zi_native id_zi_native)¶ - Parameters
[in] g: Ocean grid structure[in] gv: ocean vertical grid structure[in] param_file: Parameter file structure[inout] diag_cs: Diagnostics control structure[in] id_zl_native: ID of native layers[in] id_zi_native: ID of native interfaces
-
subroutine, public mom_diag_mediator::set_masks_for_axes(G G, diag_cs diag_cs) set_masks_for_axes sets up the 2d and 3d masks for diagnostics using the current grid recorded after calling diag_update_remap_grids()
- Parameters
[in] g: The ocean grid type.diag_cs: A pointer to a type with many variables used for diagnostics
-
subroutine
set_masks_for_axes_dsamp(G G, diag_cs diag_cs)¶ - Parameters
[in] g: The ocean grid type.diag_cs: A pointer to a type with many variables used for diagnostics
-
subroutine, public mom_diag_mediator::diag_register_area_ids(diag_cs diag_cs, id_area_t id_area_t, id_area_q id_area_q) Attaches the id of cell areas to axes groups for use with cell_measures.
- Parameters
[inout] diag_cs: Diagnostics control structure[in] id_area_t: Diag_mediator id for area of h-cells[in] id_area_q: Diag_mediator id for area of q-cells
-
subroutine, public mom_diag_mediator::register_cell_measure(G G, diag diag, Time Time) Sets a handle inside diagnostics mediator to associate 3d cell measures.
- Parameters
[in] g: Ocean grid structure[inout] diag: Regulates diagnostic output[in] time: Model time
-
subroutine, public mom_diag_mediator::diag_associate_volume_cell_measure(diag_cs diag_cs, id_h_volume id_h_volume) Attaches the id of cell volumes to axes groups for use with cell_measures.
- Parameters
[inout] diag_cs: Diagnostics control structure[in] id_h_volume: Diag_manager id for volume of h-cells
-
integer function, public mom_diag_mediator::diag_get_volume_cell_measure_dm_id(diag_cs diag_cs) Returns diag_manager id for cell measure of h-cells.
- Parameters
[in] diag_cs: Diagnostics control structure
-
subroutine, public mom_diag_mediator::define_axes_group(diag_cs diag_cs, handles handles, axes axes, nz nz, vertical_coordinate_number vertical_coordinate_number, x_cell_method x_cell_method, y_cell_method y_cell_method, v_cell_method v_cell_method, is_h_point is_h_point, is_q_point is_q_point, is_u_point is_u_point, is_v_point is_v_point, is_layer is_layer, is_interface is_interface, is_native is_native, needs_remapping needs_remapping, needs_interpolating needs_interpolating, xyave_axes xyave_axes) Defines a group of “axes” from list of handles.
- Parameters
[in] diag_cs: Diagnostics control structure[in] handles: A list of 1D axis handles[out] axes: The group of 1D axes[in] nz: Number of layers in this diagnostic grid[in] vertical_coordinate_number: Index number for vertical coordinate[in] x_cell_method: A x-direction cell method used to construct the “cell_methods” attribute in CF convention[in] y_cell_method: A y-direction cell method used to construct the “cell_methods” attribute in CF convention[in] v_cell_method: A vertical direction cell method used to construct the “cell_methods” attribute in CF convention[in] is_h_point: If true, indicates this axes group for h-point located fields[in] is_q_point: If true, indicates this axes group for q-point located fields[in] is_u_point: If true, indicates this axes group for u-point located fields[in] is_v_point: If true, indicates this axes group for v-point located fields[in] is_layer: If true, indicates that this axes group is for a layer vertically-located field.[in] is_interface: If true, indicates that this axes group is for an interface vertically-located field.[in] is_native: If true, indicates that this axes group is for a native model grid. False for any other grid.[in] needs_remapping: If true, indicates that this axes group is for a intensive layer-located field that must be remapped to these axes. Used for rank>2.[in] needs_interpolating: If true, indicates that this axes group is for a sampled interface-located field that must be interpolated to these axes. Used for rank>2.xyave_axes: The corresponding axes group for horizontally area-average diagnostics
-
subroutine
define_axes_group_dsamp(diag_cs diag_cs, handles handles, axes axes, dl dl, nz nz, vertical_coordinate_number vertical_coordinate_number, x_cell_method x_cell_method, y_cell_method y_cell_method, v_cell_method v_cell_method, is_h_point is_h_point, is_q_point is_q_point, is_u_point is_u_point, is_v_point is_v_point, is_layer is_layer, is_interface is_interface, is_native is_native, needs_remapping needs_remapping, needs_interpolating needs_interpolating, xyave_axes xyave_axes)¶ Defines a group of downsampled “axes” from list of handles.
- Parameters
[in] diag_cs: Diagnostics control structure[in] handles: A list of 1D axis handles[out] axes: The group of 1D axes[in] dl: Downsample level[in] nz: Number of layers in this diagnostic grid[in] vertical_coordinate_number: Index number for vertical coordinate[in] x_cell_method: A x-direction cell method used to construct the “cell_methods” attribute in CF convention[in] y_cell_method: A y-direction cell method used to construct the “cell_methods” attribute in CF convention[in] v_cell_method: A vertical direction cell method used to construct the “cell_methods” attribute in CF convention[in] is_h_point: If true, indicates this axes group for h-point located fields[in] is_q_point: If true, indicates this axes group for q-point located fields[in] is_u_point: If true, indicates this axes group for u-point located fields[in] is_v_point: If true, indicates this axes group for v-point located fields[in] is_layer: If true, indicates that this axes group is for a layer vertically-located field.[in] is_interface: If true, indicates that this axes group is for an interface vertically-located field.[in] is_native: If true, indicates that this axes group is for a native model grid. False for any other grid.[in] needs_remapping: If true, indicates that this axes group is for a intensive layer-located field that must be remapped to these axes. Used for rank>2.[in] needs_interpolating: If true, indicates that this axes group is for a sampled interface-located field that must be interpolated to these axes. Used for rank>2.xyave_axes: The corresponding axes group for horizontally area-average diagnostics
-
subroutine, public mom_diag_mediator::set_diag_mediator_grid(G G, diag_cs diag_cs) Set up the array extents for doing diagnostics.
- Parameters
[inout] g: The ocean’s grid structure[inout] diag_cs: Structure used to regulate diagnostic output
-
subroutine
post_data_0d(diag_field_id diag_field_id, field field, diag_cs diag_cs, is_static is_static)¶ Make a real scalar diagnostic available for averaging or output.
- Parameters
[in] diag_field_id: The id for an output variable returned by a previous call to register_diag_field.[in] field: real value being offered for output or averaging[in] diag_cs: Structure used to regulate diagnostic output[in] is_static: If true, this is a static field that is always offered.
-
subroutine, public mom_diag_mediator::post_data_1d_k(diag_field_id diag_field_id, field field, diag_cs diag_cs, is_static is_static) Make a real 1-d array diagnostic available for averaging or output.
- Parameters
[in] diag_field_id: The id for an output variable returned by a previous call to register_diag_field.[in] field: 1-d array being offered for output or averaging[in] diag_cs: Structure used to regulate diagnostic output[in] is_static: If true, this is a static field that is always offered.
-
subroutine
post_data_2d(diag_field_id diag_field_id, field field, diag_cs diag_cs, is_static is_static, mask mask)¶ Make a real 2-d array diagnostic available for averaging or output.
- Parameters
[in] diag_field_id: The id for an output variable returned by a previous call to register_diag_field.[in] field: 2-d array being offered for output or averaging[in] diag_cs: Structure used to regulate diagnostic output[in] is_static: If true, this is a static field that is always offered.[in] mask: If present, use this real array as the data mask.
-
subroutine
post_data_2d_low(diag diag, field field, diag_cs diag_cs, is_static is_static, mask mask)¶ Make a real 2-d array diagnostic available for averaging or output using a diag_type instead of an integer id.
- Parameters
[in] diag: A structure describing the diagnostic to post[in] field: 2-d array being offered for output or averaging[in] diag_cs: Structure used to regulate diagnostic output[in] is_static: If true, this is a static field that is always offered.[in] mask: If present, use this real array as the data mask.
-
subroutine
post_data_3d(diag_field_id diag_field_id, field field, diag_cs diag_cs, is_static is_static, mask mask, alt_h alt_h)¶ Make a real 3-d array diagnostic available for averaging or output.
- Parameters
[in] diag_field_id: The id for an output variable returned by a previous call to register_diag_field.[in] field: 3-d array being offered for output or averaging[in] diag_cs: Structure used to regulate diagnostic output[in] is_static: If true, this is a static field that is always offered.[in] mask: If present, use this real array as the data mask.[in] alt_h: An alternate thickness to use for vertically
-
subroutine
post_data_3d_low(diag diag, field field, diag_cs diag_cs, is_static is_static, mask mask)¶ Make a real 3-d array diagnostic available for averaging or output using a diag_type instead of an integer id.
- Parameters
[in] diag: A structure describing the diagnostic to post[in] field: 3-d array being offered for output or averaging[in] diag_cs: Structure used to regulate diagnostic output[in] is_static: If true, this is a static field that is always offered.[in] mask: If present, use this real array as the data mask.
-
subroutine
post_xy_average(diag_cs diag_cs, diag diag, field field)¶ Post the horizontally area-averaged diagnostic.
- Parameters
[in] diag: This diagnostic[in] field: Diagnostic field[in] diag_cs: Diagnostics mediator control structure
-
subroutine, public mom_diag_mediator::enable_averaging(time_int_in time_int_in, time_end_in time_end_in, diag_cs diag_cs) This subroutine enables the accumulation of time averages over the specified time interval.
- Parameters
[in] time_int_in: The time interval [s] over which any values that are offered are valid.[in] time_end_in: The end time of the valid interval[inout] diag_cs: Structure used to regulate diagnostic output
-
subroutine, public mom_diag_mediator::disable_averaging(diag_cs diag_cs) Call this subroutine to avoid averaging any offered fields.
- Parameters
[inout] diag_cs: Structure used to regulate diagnostic output
-
logical function, public mom_diag_mediator::query_averaging_enabled(diag_cs diag_cs, time_int time_int, time_end time_end) Call this subroutine to determine whether the averaging is currently enabled. .true. is returned if it is.
- Parameters
[in] diag_cs: Structure used to regulate diagnostic output[out] time_int: Current setting of diagtime_int [s][out] time_end: Current setting of diagtime_end
-
type(time_type) function, public mom_diag_mediator::get_diag_time_end(diag_cs diag_cs) This function returns the valid end time for use with diagnostics that are handled outside of the MOM6 diagnostics infrastructure.
- Parameters
[in] diag_cs: Structure used to regulate diagnostic output
-
integer function, public mom_diag_mediator::register_diag_field(module_name module_name, field_name field_name, axes_in axes_in, init_time init_time, long_name long_name, units units, missing_value missing_value, range range, mask_variant mask_variant, standard_name standard_name, verbose verbose, do_not_log do_not_log, err_msg err_msg, interp_method interp_method, tile_count tile_count, cmor_field_name cmor_field_name, cmor_long_name cmor_long_name, cmor_units cmor_units, cmor_standard_name cmor_standard_name, cell_methods cell_methods, x_cell_method x_cell_method, y_cell_method y_cell_method, v_cell_method v_cell_method, conversion conversion, v_extensive v_extensive) Returns the “diag_mediator” handle for a group (native, CMOR, z-coord, …) of diagnostics derived from one field.
- Parameters
[in] module_name: Name of this module, usually “ocean_model” or “ice_shelf_model”[in] field_name: Name of the diagnostic field[in] axes_in: Container w/ up to 3 integer handles that indicates axes for this field[in] init_time: Time at which a field is first available?[in] long_name: Long name of a field.[in] units: Units of a field.[in] standard_name: Standardized name associated with a field[in] missing_value: A value that indicates missing values.[in] range: Valid range of a variable (not used in MOM?)[in] mask_variant: If true a logical mask must be provided with post_data calls (not used in MOM?)[in] verbose: If true, FMS is verbose (not used in MOM?)[in] do_not_log: If true, do not log something (not used in MOM?)[out] err_msg: String into which an error message might be placed (not used in MOM?)[in] interp_method: If ‘none’ indicates the field should not be interpolated as a scalar[in] tile_count: no clue (not used in MOM?)[in] cmor_field_name: CMOR name of a field[in] cmor_long_name: CMOR long name of a field[in] cmor_units: CMOR units of a field[in] cmor_standard_name: CMOR standardized name associated with a field[in] cell_methods: String to append as cell_methods attribute. Use ‘’ to have no attribute. If present, this overrides the default constructed from the default for each individual axis direction.[in] x_cell_method: Specifies the cell method for the x-direction. Use ‘’ have no method.[in] y_cell_method: Specifies the cell method for the y-direction. Use ‘’ have no method.[in] v_cell_method: Specifies the cell method for the vertical direction. Use ‘’ have no method.[in] conversion: A value to multiply data by before writing to file[in] v_extensive: True for vertically extensive fields (vertically integrated). Default/absent for intensive.
-
logical function mom_diag_mediator::register_diag_field_expand_cmor(dm_id dm_id, module_name module_name, field_name field_name, axes axes, init_time init_time, long_name long_name, units units, missing_value missing_value, range range, mask_variant mask_variant, standard_name standard_name, verbose verbose, do_not_log do_not_log, err_msg err_msg, interp_method interp_method, tile_count tile_count, cmor_field_name cmor_field_name, cmor_long_name cmor_long_name, cmor_units cmor_units, cmor_standard_name cmor_standard_name, cell_methods cell_methods, x_cell_method x_cell_method, y_cell_method y_cell_method, v_cell_method v_cell_method, conversion conversion, v_extensive v_extensive) Returns True if either the native or CMOr version of the diagnostic were registered. Updates ‘dm_id’ after calling register_diag_field_expand_axes() for both native and CMOR variants of the field.
- Parameters
[inout] dm_id: The diag_mediator ID for this diagnostic group[in] module_name: Name of this module, usually “ocean_model” or “ice_shelf_model”[in] field_name: Name of the diagnostic field[in] axes: Container w/ up to 3 integer handles that indicates axes for this field[in] init_time: Time at which a field is first available?[in] long_name: Long name of a field.[in] units: Units of a field.[in] standard_name: Standardized name associated with a field[in] missing_value: A value that indicates missing values.[in] range: Valid range of a variable (not used in MOM?)[in] mask_variant: If true a logical mask must be provided with post_data calls (not used in MOM?)[in] verbose: If true, FMS is verbose (not used in MOM?)[in] do_not_log: If true, do not log something (not used in MOM?)[out] err_msg: String into which an error message might be placed (not used in MOM?)[in] interp_method: If ‘none’ indicates the field should not be interpolated as a scalar[in] tile_count: no clue (not used in MOM?)[in] cmor_field_name: CMOR name of a field[in] cmor_long_name: CMOR long name of a field[in] cmor_units: CMOR units of a field[in] cmor_standard_name: CMOR standardized name associated with a field[in] cell_methods: String to append as cell_methods attribute. Use ‘’ to have no attribute. If present, this overrides the default constructed from the default for each individual axis direction.[in] x_cell_method: Specifies the cell method for the x-direction. Use ‘’ have no method.[in] y_cell_method: Specifies the cell method for the y-direction. Use ‘’ have no method.[in] v_cell_method: Specifies the cell method for the vertical direction. Use ‘’ have no method.[in] conversion: A value to multiply data by before writing to file[in] v_extensive: True for vertically extensive fields (vertically integrated). Default/absent for intensive.
-
integer function mom_diag_mediator::register_diag_field_expand_axes(module_name module_name, field_name field_name, axes axes, init_time init_time, long_name long_name, units units, missing_value missing_value, range range, mask_variant mask_variant, standard_name standard_name, verbose verbose, do_not_log do_not_log, err_msg err_msg, interp_method interp_method, tile_count tile_count) Returns an FMS id from register_diag_field_fms (the diag_manager routine) after expanding axes (axes-group) into handles and conditionally adding an FMS area_id for cell_measures.
- Parameters
[in] module_name: Name of this module, usually “ocean_model” or “ice_shelf_model”[in] field_name: Name of the diagnostic field[in] axes: Container w/ up to 3 integer handles that indicates axes for this field[in] init_time: Time at which a field is first available?[in] long_name: Long name of a field.[in] units: Units of a field.[in] standard_name: Standardized name associated with a field[in] missing_value: A value that indicates missing values.[in] range: Valid range of a variable (not used in MOM?)[in] mask_variant: If true a logical mask must be provided with post_data calls (not used in MOM?)[in] verbose: If true, FMS is verbose (not used in MOM?)[in] do_not_log: If true, do not log something (not used in MOM?)[out] err_msg: String into which an error message might be placed (not used in MOM?)[in] interp_method: If ‘none’ indicates the field should not be interpolated as a scalar[in] tile_count: no clue (not used in MOM?)
-
subroutine
add_diag_to_list(diag_cs diag_cs, dm_id dm_id, fms_id fms_id, this_diag this_diag, axes axes, module_name module_name, field_name field_name, msg msg)¶ Create a diagnostic type and attached to list.
- Parameters
diag_cs: Diagnostics mediator control structure[inout] dm_id: The diag_mediator ID for this diagnostic group[in] fms_id: The FMS diag_manager ID for this diagnosticthis_diag: This diagnostic[in] axes: Container w/ up to 3 integer handles that indicates axes for this field[in] module_name: Name of this module, usually “ocean_model” or “ice_shelf_model”[in] field_name: Name of diagnostic[in] msg: Message for errors
-
subroutine
add_xyz_method(diag diag, axes axes, x_cell_method x_cell_method, y_cell_method y_cell_method, v_cell_method v_cell_method, v_extensive v_extensive)¶ Adds the encoded “cell_methods” for a diagnostics as a diag% property This allows access to the cell_method for a given diagnostics at the time of sending.
- Parameters
diag: This diagnostic[in] axes: Container w/ up to 3 integer handles that indicates axes for this field[in] x_cell_method: Specifies the cell method for the x-direction. Use ‘’ have no method.[in] y_cell_method: Specifies the cell method for the y-direction. Use ‘’ have no method.[in] v_cell_method: Specifies the cell method for the vertical direction. Use ‘’ have no method.[in] v_extensive: True for vertically extensive fields (vertically integrated). Default/absent for intensive.
-
subroutine
attach_cell_methods(id id, axes axes, ostring ostring, cell_methods cell_methods, x_cell_method x_cell_method, y_cell_method y_cell_method, v_cell_method v_cell_method, v_extensive v_extensive)¶ Attaches “cell_methods” attribute to a variable based on defaults for axes_grp or optional arguments.
- Parameters
[in] id: Handle to diagnostic[in] axes: Container w/ up to 3 integer handles that indicates axes for this field[out] ostring: The cell_methods strings that would appear in the file[in] cell_methods: String to append as cell_methods attribute. Use ‘’ to have no attribute. If present, this overrides the default constructed from the default for each individual axis direction.[in] x_cell_method: Specifies the cell method for the x-direction. Use ‘’ have no method.[in] y_cell_method: Specifies the cell method for the y-direction. Use ‘’ have no method.[in] v_cell_method: Specifies the cell method for the vertical direction. Use ‘’ have no method.[in] v_extensive: True for vertically extensive fields (vertically integrated). Default/absent for intensive.
-
integer function, public mom_diag_mediator::register_scalar_field(module_name module_name, field_name field_name, init_time init_time, diag_cs diag_cs, long_name long_name, units units, missing_value missing_value, range range, standard_name standard_name, do_not_log do_not_log, err_msg err_msg, interp_method interp_method, cmor_field_name cmor_field_name, cmor_long_name cmor_long_name, cmor_units cmor_units, cmor_standard_name cmor_standard_name) - Return
An integer handle for a diagnostic array.
- Parameters
[in] module_name: Name of this module, usually “ocean_model” or “ice_shelf_model”[in] field_name: Name of the diagnostic field[in] init_time: Time at which a field is first available?[inout] diag_cs: Structure used to regulate diagnostic output[in] long_name: Long name of a field.[in] units: Units of a field.[in] standard_name: Standardized name associated with a field[in] missing_value: A value that indicates missing values.[in] range: Valid range of a variable (not used in MOM?)[in] do_not_log: If true, do not log something (not used in MOM?)[out] err_msg: String into which an error message might be placed (not used in MOM?)[in] interp_method: If ‘none’ indicates the field should not be interpolated as a scalar[in] cmor_field_name: CMOR name of a field[in] cmor_long_name: CMOR long name of a field[in] cmor_units: CMOR units of a field[in] cmor_standard_name: CMOR standardized name associated with a field
-
integer function, public mom_diag_mediator::register_static_field(module_name module_name, field_name field_name, axes axes, long_name long_name, units units, missing_value missing_value, range range, mask_variant mask_variant, standard_name standard_name, do_not_log do_not_log, interp_method interp_method, tile_count tile_count, cmor_field_name cmor_field_name, cmor_long_name cmor_long_name, cmor_units cmor_units, cmor_standard_name cmor_standard_name, area area, x_cell_method x_cell_method, y_cell_method y_cell_method, area_cell_method area_cell_method, conversion conversion) Registers a static diagnostic, returning an integer handle.
- Return
An integer handle for a diagnostic array.
- Parameters
[in] module_name: Name of this module, usually “ocean_model” or “ice_shelf_model”[in] field_name: Name of the diagnostic field[in] axes: Container w/ up to 3 integer handles that indicates axes for this field[in] long_name: Long name of a field.[in] units: Units of a field.[in] standard_name: Standardized name associated with a field[in] missing_value: A value that indicates missing values.[in] range: Valid range of a variable (not used in MOM?)[in] mask_variant: If true a logical mask must be provided with post_data calls (not used in MOM?)[in] do_not_log: If true, do not log something (not used in MOM?)[in] interp_method: If ‘none’ indicates the field should not be interpolated as a scalar[in] tile_count: no clue (not used in MOM?)[in] cmor_field_name: CMOR name of a field[in] cmor_long_name: CMOR long name of a field[in] cmor_units: CMOR units of a field[in] cmor_standard_name: CMOR standardized name associated with a field[in] area: fms_id for area_t[in] x_cell_method: Specifies the cell method for the x-direction.[in] y_cell_method: Specifies the cell method for the y-direction.[in] area_cell_method: Specifies the cell method for area[in] conversion: A value to multiply data by before writing to file
-
subroutine
describe_option(opt_name opt_name, value value, diag_CS diag_CS)¶ Describe an option setting in the diagnostic files.
- Parameters
[in] opt_name: The name of the option[in] value: A character string with the setting of the option.[in] diag_cs: Structure used to regulate diagnostic output
-
integer function, public mom_diag_mediator::ocean_register_diag(var_desc var_desc, G G, diag_CS diag_CS, day day) Registers a diagnostic using the information encapsulated in the vardesc type argument and returns an integer handle to this diagostic. That integer handle is negative if the diagnostic is unused.
- Return
An integer handle to this diagnostic.
- Parameters
[in] var_desc: The vardesc type describing the diagnostic[in] g: The ocean’s grid type[in] diag_cs: The diagnotic control structure[in] day: The current model time
-
subroutine, public mom_diag_mediator::diag_mediator_infrastructure_init(err_msg err_msg) - Parameters
[out] err_msg: An error message
-
subroutine, public mom_diag_mediator::diag_mediator_init(G G, GV GV, US US, nz nz, param_file param_file, diag_cs diag_cs, doc_file_dir doc_file_dir) diag_mediator_init initializes the MOM diag_mediator and opens the available diagnostics file, if appropriate.
- Parameters
[inout] g: The ocean grid type.[in] gv: The ocean vertical grid structure[in] us: A dimensional unit scaling type[in] nz: The number of layers in the model’s native grid.[in] param_file: Parameter file structure[inout] diag_cs: A pointer to a type with many variables used for diagnostics[in] doc_file_dir: A directory in which to create the file
-
subroutine, public mom_diag_mediator::diag_set_state_ptrs(h h, T T, S S, eqn_of_state eqn_of_state, diag_cs diag_cs) Set pointers to the default state fields used to remap diagnostics.
- Parameters
[in] h: the model thickness array[in] t: the model temperature array[in] s: the model salinity array[in] eqn_of_state: Equation of state structure[inout] diag_cs: diag mediator control structure
-
subroutine, public mom_diag_mediator::diag_update_remap_grids(diag_cs diag_cs, alt_h alt_h, alt_T alt_T, alt_S alt_S) Build/update vertical grids for diagnostic remapping.
- Note
The target grids need to be updated whenever sea surface height changes.
- Parameters
[inout] diag_cs: Diagnostics control structure[in] alt_h: Used if remapped grids should be something other than the current thicknesses[in] alt_t: Used if remapped grids should be something other than the current temperatures[in] alt_s: Used if remapped grids should be something other than the current salinity
-
subroutine, public mom_diag_mediator::diag_masks_set(G G, nz nz, diag_cs diag_cs) Sets up the 2d and 3d masks for native diagnostics.
- Parameters
[in] g: The ocean grid type.[in] nz: The number of layers in the model’s native grid.diag_cs: A pointer to a type with many variables used for diagnostics
-
subroutine, public mom_diag_mediator::diag_mediator_close_registration(diag_CS diag_CS) - Parameters
[inout] diag_cs: Structure used to regulate diagnostic output
-
subroutine, public mom_diag_mediator::diag_mediator_end(time time, diag_CS diag_CS, end_diag_manager end_diag_manager) - Parameters
[in] time: The current model time[inout] diag_cs: Structure used to regulate diagnostic output[in] end_diag_manager: If true, call diag_manager_end()
-
character(len=15) function mom_diag_mediator::i2s(a a, n_in n_in) Convert the first n elements (up to 3) of an integer array to an underscore delimited string.
- Return
The returned string
- Parameters
[in] a: The array of integers to translate[in] n_in: The number of elements to translate, by default all
-
integer function mom_diag_mediator::get_new_diag_id(diag_cs diag_cs) Returns a new diagnostic id, it may be necessary to expand the diagnostics array.
- Parameters
[inout] diag_cs: Diagnostics control structure
-
subroutine
initialize_diag_type(diag diag)¶ Initializes a diag_type (used after allocating new memory)
- Parameters
[inout] diag: diag_type to be initialized
-
subroutine
alloc_diag_with_id(diag_id diag_id, diag_cs diag_cs, diag diag)¶ Make a new diagnostic. Either use memory which is in the array of ‘primary’ diagnostics, or if that is in use, insert it to the list of secondary diags.
- Parameters
[in] diag_id: id for the diagnostic[inout] diag_cs: structure used to regulate diagnostic outputdiag: structure representing a diagnostic (inout)
-
subroutine
log_available_diag(used used, module_name module_name, field_name field_name, cell_methods_string cell_methods_string, comment comment, diag_CS diag_CS, long_name long_name, units units, standard_name standard_name)¶ Log a diagnostic to the available diagnostics file.
- Parameters
[in] used: Whether this diagnostic was in the diag_table or not[in] module_name: Name of the diagnostic module[in] field_name: Name of this diagnostic field[in] cell_methods_string: The spatial component of the CF cell_methods attribute[in] comment: A comment to append after [Used|Unused][in] diag_cs: The diagnotics control structure[in] long_name: CF long name of diagnostic[in] units: Units for diagnostic[in] standard_name: CF standardized name of diagnostic
-
subroutine
log_chksum_diag(docunit docunit, description description, chksum chksum)¶ Log the diagnostic chksum to the chksum diag file.
- Parameters
[in] docunit: Handle of the log file[in] description: Name of the diagnostic module[in] chksum: chksum of the diagnostic
-
subroutine, public mom_diag_mediator::diag_grid_storage_init(grid_storage grid_storage, G G, diag diag) Allocates fields necessary to store diagnostic remapping fields.
- Parameters
[inout] grid_storage: Structure containing a snapshot of the target grids[in] g: Horizontal grid[in] diag: Diagnostic control structure used as the contructor template for this routine
-
subroutine, public mom_diag_mediator::diag_copy_diag_to_storage(grid_storage grid_storage, h_state h_state, diag diag) Copy from the main diagnostic arrays to the grid storage as well as the native thicknesses.
- Parameters
[inout] grid_storage: Structure containing a snapshot of the target grids[in] h_state: Current model thicknesses[in] diag: Diagnostic control structure used as the contructor
-
subroutine, public mom_diag_mediator::diag_copy_storage_to_diag(diag diag, grid_storage grid_storage) Copy from the stored diagnostic arrays to the main diagnostic grids.
- Parameters
[inout] diag: Diagnostic control structure used as the contructor[in] grid_storage: Structure containing a snapshot of the target grids
-
subroutine, public mom_diag_mediator::diag_save_grids(diag diag) Save the current diagnostic grids in the temporary structure within diag.
- Parameters
[inout] diag: Diagnostic control structure used as the contructor
-
subroutine, public mom_diag_mediator::diag_restore_grids(diag diag) Restore the diagnostic grids from the temporary structure within diag.
- Parameters
[inout] diag: Diagnostic control structure used as the contructor
-
subroutine, public mom_diag_mediator::diag_grid_storage_end(grid_storage grid_storage) Deallocates the fields in the remapping fields container.
- Parameters
[inout] grid_storage: Structure containing a snapshot of the target grids
-
subroutine
downsample_diag_masks_set(G G, nz nz, diag_cs diag_cs)¶ - Parameters
[in] g: The ocean grid type.[in] nz: The number of layers in the model’s native grid.diag_cs: A pointer to a type with many variables used for diagnostics
-
subroutine
downsample_diag_indices_get(fo1 fo1, fo2 fo2, dl dl, diag_cs diag_cs, isv isv, iev iev, jsv jsv, jev jev)¶ Get the diagnostics-compute indices (to be passed to send_data) based on the shape of the diag field (the same way they are deduced for non-downsampled fields)
- Parameters
[in] fo1: The size of the diag field in x[in] fo2: The size of the diag field in y[in] dl: Integer downsample level[in] diag_cs: Structure used to regulate diagnostic output[out] isv: i-start index for diagnostics[out] iev: i-end index for diagnostics[out] jsv: j-start index for diagnostics[out] jev: j-end index for diagnostics
-
subroutine
downsample_diag_field_3d(locfield locfield, locfield_dsamp locfield_dsamp, dl dl, diag_cs diag_cs, diag diag, isv isv, iev iev, jsv jsv, jev jev, mask mask)¶ This subroutine allocates and computes a downsampled array from an input array It also determines the diagnostics-compurte indices for the downsampled array 3d interface.
- Parameters
locfield: Input array pointer[inout] locfield_dsamp: Output (downsampled) array[in] diag_cs: Structure used to regulate diagnostic output[in] diag: A structure describing the diagnostic to post[in] dl: Level of down sampling[inout] isv: i-start index for diagnostics[inout] iev: i-end index for diagnostics[inout] jsv: j-start index for diagnostics[inout] jev: j-end index for diagnostics[in] mask: If present, use this real array as the data mask.
-
subroutine
downsample_diag_field_2d(locfield locfield, locfield_dsamp locfield_dsamp, dl dl, diag_cs diag_cs, diag diag, isv isv, iev iev, jsv jsv, jev jev, mask mask)¶ This subroutine allocates and computes a downsampled array from an input array It also determines the diagnostics-compurte indices for the downsampled array 2d interface.
- Parameters
locfield: Input array pointer[inout] locfield_dsamp: Output (downsampled) array[in] diag_cs: Structure used to regulate diagnostic output[in] diag: A structure describing the diagnostic to post[in] dl: Level of down sampling[inout] isv: i-start index for diagnostics[inout] iev: i-end index for diagnostics[inout] jsv: j-start index for diagnostics[inout] jev: j-end index for diagnostics[in] mask: If present, use this real array as the data mask.
-
subroutine
downsample_field_3d(field_in field_in, field_out field_out, dl dl, method method, mask mask, diag_cs diag_cs, diag diag, isv_o isv_o, jsv_o jsv_o, isv_d isv_d, iev_d iev_d, jsv_d jsv_d, jev_d jev_d)¶ This subroutine allocates and computes a down sampled 3d array given an input array The down sample method is based on the “cell_methods” for the diagnostics as explained in the above table.
-
subroutine
downsample_field_2d(field_in field_in, field_out field_out, dl dl, method method, mask mask, diag_cs diag_cs, diag diag, isv_o isv_o, jsv_o jsv_o, isv_d isv_d, iev_d iev_d, jsv_d jsv_d, jev_d jev_d)¶ This subroutine allocates and computes a down sampled 2d array given an input array The down sample method is based on the “cell_methods” for the diagnostics as explained in the above table.
- Parameters
field_in: Original field to be down sampledfield_out: Down sampled field[in] dl: Level of down sampling[in] method: Sampling methodmask: Mask for field[in] diag_cs: Structure used to regulate diagnostic output[in] diag: A structure describing the diagnostic to post[in] isv_o: Original i-start index[in] jsv_o: Original j-start index[in] isv_d: i-start index of down sampled data[in] iev_d: i-end index of down sampled data[in] jsv_d: j-start index of down sampled data[in] jev_d: j-end index of down sampled data
-
subroutine
downsample_mask_2d(field_in field_in, field_out field_out, dl dl, isc_o isc_o, jsc_o jsc_o, isc_d isc_d, iec_d iec_d, jsc_d jsc_d, jec_d jec_d, isd_d isd_d, ied_d ied_d, jsd_d jsd_d, jed_d jed_d)¶ Allocate and compute the 2d down sampled mask The masks are down sampled based on a minority rule, i.e., a coarse cell is open (1) if at least one of the sub-cells are open, otherwise it’s closed (0)
- Parameters
[in] field_in: Original field to be down sampledfield_out: Down sampled field[in] dl: Level of down sampling[in] isc_o: Original i-start index[in] jsc_o: Original j-start index[in] isc_d: Computational i-start index of down sampled data[in] iec_d: Computational i-end index of down sampled data[in] jsc_d: Computational j-start index of down sampled data[in] jec_d: Computational j-end index of down sampled data[in] isd_d: Computational i-start index of down sampled data[in] ied_d: Computational i-end index of down sampled data[in] jsd_d: Computational j-start index of down sampled data[in] jed_d: Computational j-end index of down sampled data
-
subroutine
downsample_mask_3d(field_in field_in, field_out field_out, dl dl, isc_o isc_o, jsc_o jsc_o, isc_d isc_d, iec_d iec_d, jsc_d jsc_d, jec_d jec_d, isd_d isd_d, ied_d ied_d, jsd_d jsd_d, jed_d jed_d)¶ Allocate and compute the 3d down sampled mask The masks are down sampled based on a minority rule, i.e., a coarse cell is open (1) if at least one of the sub-cells are open, otherwise it’s closed (0)
- Parameters
[in] field_in: Original field to be down sampledfield_out: down sampled field[in] dl: Level of down sampling[in] isc_o: Original i-start index[in] jsc_o: Original j-start index[in] isc_d: Computational i-start index of down sampled data[in] iec_d: Computational i-end index of down sampled data[in] jsc_d: Computational j-start index of down sampled data[in] jec_d: Computational j-end index of down sampled data[in] isd_d: Computational i-start index of down sampled data[in] ied_d: Computational i-end index of down sampled data[in] jsd_d: Computational j-start index of down sampled data[in] jed_d: Computational j-end index of down sampled data
Variables
-
integer
psp= 121¶ x:point,y:sum,z:point
-
integer
pss= 122¶ x:point,y:sum,z:point
-
integer
psm= 123¶ x:point,y:sum,z:mean
-
integer
pmp= 131¶ x:point,y:mean,z:point
-
integer
pmm= 133¶ x:point,y:mean,z:mean
-
integer
spp= 211¶ x:sum,y:point,z:point
-
integer
sps= 212¶ x:sum,y:point,z:sum
-
integer
ssp= 221¶ x:sum;y:sum,z:point
-
integer
mpp= 311¶ x:mean,y:point,z:point
-
integer
mpm= 313¶ x:mean,y:point,z:mean
-
integer
mmp= 331¶ x:mean,y:mean,z:point
-
integer
mms= 332¶ x:mean,y:mean,z:sum
-
integer
sss= 222¶ x:sum,y:sum,z:sum
-
integer
mmm= 333¶ x:mean,y:mean,z:mean
-
integer
msk= -1¶ Use the downsample method of a mask.
-
integer
-
namespace
mom_diag_remap¶ provides runtime remapping of diagnostics to z star, sigma and rho vertical coordinates.
The diag_remap_ctrl type represents a remapping of diagnostics to a particular vertical coordinate. The module is used by the diag mediator module in the following way:
diag_remap_init() is called to initialize a diag_remap_ctrl instance.
diag_remap_configure_axes() is called to read the configuration file and set up the vertical coordinate / axes definitions.
diag_remap_get_axes_info() returns information needed for the diag mediator to define new axes for the remapped diagnostics.
diag_remap_update() is called periodically (whenever h, T or S change) to either create or update the target remapping grids.
diag_remap_do_remap() is called from within a diag post() to do the remapping before the diagnostic is written out.
Functions
-
subroutine, public mom_diag_remap::diag_remap_init(remap_cs remap_cs, coord_tuple coord_tuple) Initialize a diagnostic remapping type with the given vertical coordinate.
- Parameters
[inout] remap_cs: Diag remapping control structure[in] coord_tuple: A string in form of MODULE_SUFFIX PARAMETER_SUFFIX COORDINATE_NAME
-
subroutine, public mom_diag_remap::diag_remap_end(remap_cs remap_cs) De-init a diagnostic remapping type. Free allocated memory.
- Parameters
[inout] remap_cs: Diag remapping control structure
-
subroutine, public mom_diag_remap::diag_remap_diag_registration_closed(remap_cs remap_cs) Inform that all diagnostics have been registered. If _set_active() has not been called on the remapping control structure will be disabled. This saves time in the case that a vertical coordinate was configured but no diagnostics which use the coordinate appeared in the diag_table.
- Parameters
[inout] remap_cs: Diag remapping control structure
-
subroutine, public mom_diag_remap::diag_remap_set_active(remap_cs remap_cs) Indicate that this remapping type is actually used by the diag manager. If this is never called then the type will be disabled to save time. See further explanation with diag_remap_registration_closed.
- Parameters
[inout] remap_cs: Diag remapping control structure
-
subroutine, public mom_diag_remap::diag_remap_configure_axes(remap_cs remap_cs, GV GV, US US, param_file param_file) Configure the vertical axes for a diagnostic remapping control structure. Reads a configuration parameters to determine coordinate generation.
- Parameters
[inout] remap_cs: Diag remap control structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: Parameter file structure
-
subroutine, public mom_diag_remap::diag_remap_get_axes_info(remap_cs remap_cs, nz nz, id_layer id_layer, id_interface id_interface) Get layer and interface axes ids for this coordinate Needed when defining axes groups.
- Parameters
[in] remap_cs: Diagnostic coordinate control structure[out] nz: Number of vertical levels for the coordinate[out] id_layer: 1D-axes id for layer points[out] id_interface: 1D-axes id for interface points
-
logical function, public mom_diag_remap::diag_remap_axes_configured(remap_cs remap_cs) Whether or not the axes for this vertical coordinated has been configured. Configuration is complete when diag_remap_configure_axes() has been successfully called.
- Parameters
[in] remap_cs: Diagnostic coordinate control structure
-
subroutine, public mom_diag_remap::diag_remap_update(remap_cs remap_cs, G G, GV GV, US US, h h, T T, S S, eqn_of_state eqn_of_state) Build/update target vertical grids for diagnostic remapping.
- Note
The target grids need to be updated whenever sea surface height or layer thicknesses changes. In the case of density-based coordinates then technically we should also regenerate the target grid whenever T/S change.
- Parameters
[inout] remap_cs: Diagnostic coordinate control structureg: The ocean’s grid type[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[in] h: New thickness[in] t: New T[in] s: New Seqn_of_state: A pointer to the equation of state
-
subroutine, public mom_diag_remap::diag_remap_do_remap(remap_cs remap_cs, G G, GV GV, h h, staggered_in_x staggered_in_x, staggered_in_y staggered_in_y, mask mask, missing_value missing_value, field field, remapped_field remapped_field) Remap diagnostic field to alternative vertical grid.
- Parameters
[in] remap_cs: Diagnostic coodinate control structure[in] g: Ocean grid structure[in] gv: ocean vertical grid structure[in] h: The current thicknesses[in] staggered_in_x: True is the x-axis location is at u or q points[in] staggered_in_y: True is the y-axis location is at v or q pointsmask: A mask for the field[in] missing_value: A missing_value to assign land/vanished points[in] field: The diagnostic field to be remapped[inout] remapped_field: Field remapped to new coordinate
-
subroutine, public mom_diag_remap::diag_remap_calc_hmask(remap_cs remap_cs, G G, mask mask) Calculate masks for target grid.
- Parameters
[in] remap_cs: Diagnostic coodinate control structure[in] g: Ocean grid structure[out] mask: h-point mask for target grid
-
subroutine, public mom_diag_remap::vertically_reintegrate_diag_field(remap_cs remap_cs, G G, h h, staggered_in_x staggered_in_x, staggered_in_y staggered_in_y, mask mask, missing_value missing_value, field field, reintegrated_field reintegrated_field) Vertically re-grid an already vertically-integrated diagnostic field to alternative vertical grid.
- Parameters
[in] remap_cs: Diagnostic coodinate control structure[in] g: Ocean grid structure[in] h: The current thicknesses[in] staggered_in_x: True is the x-axis location is at u or q points[in] staggered_in_y: True is the y-axis location is at v or q pointsmask: A mask for the field[in] missing_value: A missing_value to assign land/vanished points[in] field: The diagnostic field to be remapped[inout] reintegrated_field: Field argument remapped to alternative coordinate
-
subroutine, public mom_diag_remap::vertically_interpolate_diag_field(remap_cs remap_cs, G G, h h, staggered_in_x staggered_in_x, staggered_in_y staggered_in_y, mask mask, missing_value missing_value, field field, interpolated_field interpolated_field) Vertically interpolate diagnostic field to alternative vertical grid.
- Parameters
[in] remap_cs: Diagnostic coodinate control structure[in] g: Ocean grid structure[in] h: The current thicknesses[in] staggered_in_x: True is the x-axis location is at u or q points[in] staggered_in_y: True is the y-axis location is at v or q pointsmask: A mask for the field[in] missing_value: A missing_value to assign land/vanished points[in] field: The diagnostic field to be remapped[inout] interpolated_field: Field argument remapped to alternative coordinate
-
subroutine, public mom_diag_remap::horizontally_average_diag_field(G G, h h, staggered_in_x staggered_in_x, staggered_in_y staggered_in_y, is_layer is_layer, is_extensive is_extensive, missing_value missing_value, field field, averaged_field averaged_field, averaged_mask averaged_mask) Horizontally average field.
- Parameters
[in] g: Ocean grid structure[in] h: The current thicknesses[in] staggered_in_x: True if the x-axis location is at u or q points[in] staggered_in_y: True if the y-axis location is at v or q points[in] is_layer: True if the z-axis location is at h points[in] is_extensive: True if the z-direction is spatially integrated (over layers)[in] missing_value: A missing_value to assign land/vanished points[in] field: The diagnostic field to be remapped[inout] averaged_field: Field argument horizontally averaged[inout] averaged_mask: Mask for horizontally averaged field
-
namespace
mom_diag_vkernels¶ Provides kernels for single-column interpolation, re-integration (re-mapping of integrated quantities) and intensive-variable remapping in the vertical.
Functions
-
subroutine, public mom_diag_vkernels::interpolate_column(nsrc nsrc, h_src h_src, u_src u_src, ndest ndest, h_dest h_dest, missing_value missing_value, u_dest u_dest) Linearly interpolate interface data, u_src, from grid h_src to a grid h_dest.
- Parameters
[in] nsrc: Number of source cells[in] h_src: Thickness of source cells[in] u_src: Values at source cell interfaces[in] ndest: Number of destination cells[in] h_dest: Thickness of destination cells[in] missing_value: Value to assign in vanished cells[inout] u_dest: Interpolated value at destination cell interfaces
-
subroutine, public mom_diag_vkernels::reintegrate_column(nsrc nsrc, h_src h_src, uh_src uh_src, ndest ndest, h_dest h_dest, missing_value missing_value, uh_dest uh_dest) Conservatively calculate integrated data, uh_dest, on grid h_dest, from layer-integrated data, uh_src, on grid h_src.
- Parameters
[in] nsrc: Number of source cells[in] h_src: Thickness of source cells[in] uh_src: Values at source cell interfaces[in] ndest: Number of destination cells[in] h_dest: Thickness of destination cells[in] missing_value: Value to assign in vanished cells[inout] uh_dest: Interpolated value at destination cell interfaces
-
logical function, public mom_diag_vkernels::diag_vkernels_unit_tests(verbose verbose) Returns true if any unit tests for module MOM_diag_vkernels fail.
- Parameters
[in] verbose: If true, write results to stdout
-
logical function mom_diag_vkernels::test_interp(verbose verbose, missing_value missing_value, msg msg, nsrc nsrc, h_src h_src, u_src u_src, ndest ndest, h_dest h_dest, u_true u_true) Returns true if a test of interpolate_column() produces the wrong answer.
- Parameters
[in] verbose: If true, write results to stdout[in] missing_value: Value to indicate missing data[in] msg: Message to label test[in] nsrc: Number of source cells[in] h_src: Thickness of source cells[in] u_src: Values at source cell interfaces[in] ndest: Number of destination cells[in] h_dest: Thickness of destination cells[in] u_true: Correct value at destination cell interfaces
-
logical function mom_diag_vkernels::test_reintegrate(verbose verbose, missing_value missing_value, msg msg, nsrc nsrc, h_src h_src, uh_src uh_src, ndest ndest, h_dest h_dest, uh_true uh_true) Returns true if a test of reintegrate_column() produces the wrong answer.
- Parameters
[in] verbose: If true, write results to stdout[in] missing_value: Value to indicate missing data[in] msg: Message to label test[in] nsrc: Number of source cells[in] h_src: Thickness of source cells[in] uh_src: Values of source cell stuff[in] ndest: Number of destination cells[in] h_dest: Thickness of destination cells[in] uh_true: Correct value of destination cell stuff
-
-
namespace
mom_diagnostics¶ Calculates any requested diagnostic quantities that are not calculated in the various subroutines. Diagnostic quantities are requested by allocating them memory.
Unnamed Group
-
subroutine, public mom_diagnostics::calculate_diagnostic_fields(u u, v v, h h, uh uh, vh vh, tv tv, ADp ADp, CDp CDp, p_surf p_surf, dt dt, diag_pre_sync diag_pre_sync, G G, GV GV, US US, CS CS, eta_bt eta_bt) Diagnostics not more naturally calculated elsewhere are computed here.
- Parameters
[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] u: The zonal velocity [m s-1].[in] v: The meridional velocity [m s-1].[in] h: Layer thicknesses [H ~> m or kg m-2].[in] uh: Transport through zonal faces = u*h*dy,[in] vh: Transport through meridional faces = v*h*dx,[in] tv: A structure pointing to various thermodynamic variables.[in] adp: structure with pointers to accelerations in momentum equation.[in] cdp: structure with pointers to terms in continuity equation.p_surf: A pointer to the surface pressure [Pa]. If p_surf is not associated, it is the same as setting the surface pressure to 0.[in] dt: The time difference since the last call to this subroutine [s].[in] diag_pre_sync: Target grids from previous timestep[inout] cs: Control structure returned by a previous call to diagnostics_init.[in] eta_bt: An optional barotropic
-
subroutine
find_weights(Rlist Rlist, R_in R_in, k k, nz nz, wt wt, wt_p wt_p)¶ This subroutine finds the location of R_in in an increasing ordered list, Rlist, returning as k the element such that Rlist(k) <= R_in < Rlist(k+1), and where wt and wt_p are the linear weights that should be assigned to elements k and k+1.
- Parameters
[in] rlist: The list of target densities [kg m-3][in] r_in: The density being inserted into Rlist [kg m-3][inout] k: The value of k such that Rlist(k) <= R_in < Rlist(k+1) The input value is a first guess[in] nz: The number of layers in Rlist[out] wt: The weight of layer k for interpolation, nondim[out] wt_p: The weight of layer k+1 for interpolation, nondim
-
subroutine
calculate_vertical_integrals(h h, tv tv, p_surf p_surf, G G, GV GV, US US, CS CS)¶ This subroutine calculates vertical integrals of several tracers, along with the mass-weight of these tracers, the total column mass, and the carefully calculated column height.
- Parameters
[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] h: Layer thicknesses [H ~> m or kg m-2].[in] tv: A structure pointing to various thermodynamic variables.p_surf: A pointer to the surface pressure [Pa]. If p_surf is not associated, it is the same as setting the surface pressure to 0.[inout] cs: Control structure returned by a previous call to diagnostics_init.
-
subroutine
calculate_energy_diagnostics(u u, v v, h h, uh uh, vh vh, ADp ADp, CDp CDp, G G, GV GV, US US, CS CS)¶ This subroutine calculates terms in the mechanical energy budget.
- Parameters
[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] u: The zonal velocity [m s-1].[in] v: The meridional velocity [m s-1].[in] h: Layer thicknesses [H ~> m or kg m-2].[in] uh: Transport through zonal faces=u*h*dy,[in] vh: Transport through merid faces=v*h*dx,[in] adp: Structure pointing to accelerations in momentum equation.[in] cdp: Structure pointing to terms in continuity equations.[in] us: A dimensional unit scaling type[inout] cs: Control structure returned by a previous call to diagnostics_init.
-
subroutine, public mom_diagnostics::register_time_deriv(lb lb, f_ptr f_ptr, deriv_ptr deriv_ptr, CS CS) This subroutine registers fields to calculate a diagnostic time derivative.
- Parameters
[in] lb: Lower index bound of f_ptrf_ptr: Time derivative operandderiv_ptr: Time derivative of f_ptrcs: Control structure returned by previous call to diagnostics_init.
-
subroutine
calculate_derivs(dt dt, G G, CS CS)¶ This subroutine calculates all registered time derivatives.
- Parameters
[in] dt: The time interval over which differences occur [s].[inout] g: The ocean’s grid structure.[inout] cs: Control structure returned by previous call to diagnostics_init.
-
subroutine, public mom_diagnostics::post_surface_dyn_diags(IDs IDs, G G, diag diag, sfc_state sfc_state, ssh ssh) This routine posts diagnostics of various dynamic ocean surface quantities, including velocities, speed and sea surface height, at the time the ocean state is reported back to the caller.
- Parameters
[in] ids: A structure with the diagnostic IDs.[in] g: ocean grid structure[in] diag: regulates diagnostic output[in] sfc_state: structure describing the ocean surface state[in] ssh: Time mean surface height without corrections for ice displacement [m]
-
subroutine, public mom_diagnostics::post_surface_thermo_diags(IDs IDs, G G, GV GV, US US, diag diag, dt_int dt_int, sfc_state sfc_state, tv tv, ssh ssh, ssh_ibc ssh_ibc) This routine posts diagnostics of various ocean surface and integrated quantities at the time the ocean state is reported back to the caller.
- Parameters
[in] ids: A structure with the diagnostic IDs.[in] g: ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[in] diag: regulates diagnostic output[in] dt_int: total time step associated with these diagnostics [s].[in] sfc_state: structure describing the ocean surface state[in] tv: A structure pointing to various thermodynamic variables[in] ssh: Time mean surface height without corrections for ice displacement [m][in] ssh_ibc: Time mean surface height with corrections for ice displacement and the inverse barometer [m]
-
subroutine, public mom_diagnostics::post_transport_diagnostics(G G, GV GV, uhtr uhtr, vhtr vhtr, h h, IDs IDs, diag_pre_dyn diag_pre_dyn, diag diag, dt_trans dt_trans, Reg Reg) This routine posts diagnostics of the transports, including the subgridscale contributions.
- Parameters
[inout] g: ocean grid structure[in] gv: ocean vertical grid structure[in] uhtr: Accumulated zonal thickness fluxes used to advect tracers [H m2 ~> m3 or kg][in] vhtr: Accumulated meridional thickness fluxes used to advect tracers [H m2 ~> m3 or kg][in] h: The updated layer thicknesses [H ~> m or kg m-2][in] ids: A structure with the diagnostic IDs.[inout] diag_pre_dyn: Stored grids from before dynamics[inout] diag: regulates diagnostic output[in] dt_trans: total time step associated with the transports [s].reg: Pointer to the tracer registry
-
subroutine, public mom_diagnostics::mom_diagnostics_init(MIS MIS, ADp ADp, CDp CDp, Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS, tv tv) This subroutine registers various diagnostics and allocates space for fields that other diagnostis depend upon.
- Parameters
[in] mis: For “MOM Internal State” a set of pointers to the fields and accelerations that make up the ocean’s internal physical state.[inout] adp: Structure with pointers to momentum equation terms.[inout] cdp: Structure with pointers to continuity equation terms.[in] time: Current model time.[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters.[inout] diag: Structure to regulate diagnostic output.cs: Pointer set to point to control structure for this module.[in] tv: A structure pointing to various thermodynamic variables.
-
subroutine, public mom_diagnostics::register_surface_diags(Time Time, G G, IDs IDs, diag diag, tv tv) Register diagnostics of the surface state and integrated quantities.
- Parameters
[in] time: current model time[in] g: ocean grid structure[inout] ids: A structure with the diagnostic IDs.[inout] diag: regulates diagnostic output[in] tv: A structure pointing to various thermodynamic variables
-
subroutine, public mom_diagnostics::register_transport_diags(Time Time, G G, GV GV, IDs IDs, diag diag) Register certain diagnostics related to transports.
- Parameters
[in] time: current model time[in] g: ocean grid structure[in] gv: ocean vertical grid structure[inout] ids: A structure with the diagnostic IDs.[inout] diag: regulates diagnostic output
-
subroutine, public mom_diagnostics::write_static_fields(G G, GV GV, US US, tv tv, diag diag) Offers the static fields in the ocean grid type for output via the diag_manager.
- Parameters
[in] g: ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[in] tv: A structure pointing to various thermodynamic variables[inout] diag: regulates diagnostic output
-
subroutine
set_dependent_diagnostics(MIS MIS, ADp ADp, CDp CDp, G G, CS CS)¶ This subroutine sets up diagnostics upon which other diagnostics depend.
- Parameters
[in] mis: For “MOM Internal State” a set of pointers to the fields and accelerations making up ocean internal physical state.[inout] adp: Structure pointing to accelerations in momentum equation.[inout] cdp: Structure pointing to terms in continuity equation.[in] g: The ocean’s grid structure.cs: Pointer to the control structure for this module.
-
subroutine, public mom_diagnostics::mom_diagnostics_end(CS CS, ADp ADp) Deallocate memory associated with the diagnostics module.
- Parameters
cs: Control structure returned by a previous call to diagnostics_init.[inout] adp: structure with pointers to accelerations in momentum equation.
-
-
namespace
mom_diapyc_energy_req¶ Calculates the energy requirements of mixing.
Unnamed Group
-
subroutine, public mom_diapyc_energy_req::diapyc_energy_req_test(h_3d h_3d, dt dt, tv tv, G G, GV GV, US US, CS CS, Kd_int Kd_int) This subroutine helps test the accuracy of the diapycnal mixing energy requirement code by writing diagnostics, possibly using an intensely mixing test profile of diffusivity.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] h_3d: Layer thickness before entrainment [H ~> m or kg m-2].[inout] tv: A structure containing pointers to any available thermodynamic fields. Absent fields have NULL ptrs.[in] dt: The amount of time covered by this call [T ~> s].cs: This module’s control structure.[in] kd_int: Interface diffusivities [Z2 T-1 ~> m2 s-1].
-
subroutine, public mom_diapyc_energy_req::diapyc_energy_req_calc(h_in h_in, T_in T_in, S_in S_in, Kd Kd, energy_Kd energy_Kd, dt dt, tv tv, G G, GV GV, US US, may_print may_print, CS CS) This subroutine uses a substantially refactored tridiagonal equation for diapycnal mixing of temperature and salinity to estimate the potential energy change due to diapycnal mixing in a column of water. It does this estimate 4 different ways, all of which should be equivalent, but reports only one. The various estimates are taken because they will later be used as templates for other bits of code.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] h_in: Layer thickness before entrainment, [H ~> m or kg m-2].[in] t_in: The layer temperatures [degC].[in] s_in: The layer salinities [ppt].[in] kd: The interfaces diapycnal diffusivities [Z2 T-1 ~> m2 s-1].[in] dt: The amount of time covered by this call [T ~> s].[out] energy_kd: The column-integrated rate of energy consumption by diapycnal diffusion [W m-2].[inout] tv: A structure containing pointers to any available thermodynamic fields. Absent fields have NULL ptrs.[in] may_print: If present and true, write out diagnostics of energy use.cs: This module’s control structure.
-
subroutine
find_pe_chg(Kddt_h0 Kddt_h0, dKddt_h dKddt_h, hp_a hp_a, hp_b hp_b, Th_a Th_a, Sh_a Sh_a, Th_b Th_b, Sh_b Sh_b, dT_to_dPE_a dT_to_dPE_a, dS_to_dPE_a dS_to_dPE_a, dT_to_dPE_b dT_to_dPE_b, dS_to_dPE_b dS_to_dPE_b, pres_Z pres_Z, dT_to_dColHt_a dT_to_dColHt_a, dS_to_dColHt_a dS_to_dColHt_a, dT_to_dColHt_b dT_to_dColHt_b, dS_to_dColHt_b dS_to_dColHt_b, PE_chg PE_chg, dPEc_dKd dPEc_dKd, dPE_max dPE_max, dPEc_dKd_0 dPEc_dKd_0, ColHt_cor ColHt_cor)¶ This subroutine calculates the change in potential energy and or derivatives for several changes in an interfaces’s diapycnal diffusivity times a timestep.
- Parameters
[in] kddt_h0: The previously used diffusivity at an interface times the time step and divided by the average of the thicknesses around the interface [H ~> m or kg m-2].[in] dkddt_h: The trial change in the diffusivity at an interface times the time step and divided by the average of the thicknesses around the interface [H ~> m or kg m-2].[in] hp_a: The effective pivot thickness of the layer above the interface, given by h_k plus a term that is a fraction (determined from the tridiagonal solver) of Kddt_h for the interface above [H ~> m or kg m-2].[in] hp_b: The effective pivot thickness of the layer below the interface, given by h_k plus a term that is a fraction (determined from the tridiagonal solver) of Kddt_h for the interface above [H ~> m or kg m-2].[in] th_a: An effective temperature times a thickness in the layer above, including implicit mixing effects with other yet higher layers [degC H ~> degC m or degC kg m-2].[in] sh_a: An effective salinity times a thickness in the layer above, including implicit mixing effects with other yet higher layers [ppt H ~> ppt m or ppt kg m-2].[in] th_b: An effective temperature times a thickness in the layer below, including implicit mixing effects with other yet lower layers [degC H ~> degC m or degC kg m-2].[in] sh_b: An effective salinity times a thickness in the layer below, including implicit mixing effects with other yet lower layers [ppt H ~> ppt m or ppt kg m-2].[in] dt_to_dpe_a: A factor (pres_lay*mass_lay*dSpec_vol/dT) relating a layer’s temperature change to the change in column potential energy, including all implicit diffusive changes in the temperatures of all the layers above [J m-2 degC-1].[in] ds_to_dpe_a: A factor (pres_lay*mass_lay*dSpec_vol/dS) relating a layer’s salinity change to the change in column potential energy, including all implicit diffusive changes in the salinities of all the layers above [J m-2 ppt-1].[in] dt_to_dpe_b: A factor (pres_lay*mass_lay*dSpec_vol/dT) relating a layer’s temperature change to the change in column potential energy, including all implicit diffusive changes in the temperatures of all the layers below [J m-2 degC-1].[in] ds_to_dpe_b: A factor (pres_lay*mass_lay*dSpec_vol/dS) relating a layer’s salinity change to the change in column potential energy, including all implicit diffusive changes in the salinities of all the layers below [J m-2 ppt-1].[in] pres_z: The hydrostatic interface pressure, which is used to relate the changes in column thickness to the energy that is radiated as gravity waves and unavailable to drive mixing [J m-2 Z-1 ~> J m-3].[in] dt_to_dcolht_a: A factor (mass_lay*dSColHtc_vol/dT) relating a layer’s temperature change to the change in column height, including all implicit diffusive changes in the temperatures of all the layers above [Z degC-1 ~> m degC-1].[in] ds_to_dcolht_a: A factor (mass_lay*dSColHtc_vol/dS) relating a layer’s salinity change to the change in column height, including all implicit diffusive changes in the salinities of all the layers above [Z ppt-1 ~> m ppt-1].[in] dt_to_dcolht_b: A factor (mass_lay*dSColHtc_vol/dT) relating a layer’s temperature change to the change in column height, including all implicit diffusive changes in the temperatures of all the layers below [Z degC-1 ~> m degC-1].[in] ds_to_dcolht_b: A factor (mass_lay*dSColHtc_vol/dS) relating a layer’s salinity change to the change in column height, including all implicit diffusive changes in the salinities of all the layers below [Z ppt-1 ~> m ppt-1].[out] pe_chg: The change in column potential energy from applying Kddt_h at the present interface [J m-2].[out] dpec_dkd: The partial derivative of PE_chg with Kddt_h, [J m-2 H-1 ~> J m-3 or J kg-1].[out] dpe_max: The maximum change in column potential energy that could be realizedd by applying a huge value of Kddt_h at the present interface [J m-2].[out] dpec_dkd_0: The partial derivative of PE_chg with Kddt_h in the limit where Kddt_h = 0 [J m-2 H-1 ~> J m-3 or J kg-1].[out] colht_cor: The correction to PE_chg that is made due to a net change in the column height [J m-2].
-
subroutine
find_pe_chg_orig(Kddt_h Kddt_h, h_k h_k, b_den_1 b_den_1, dTe_term dTe_term, dSe_term dSe_term, dT_km1_t2 dT_km1_t2, dS_km1_t2 dS_km1_t2, dT_to_dPE_k dT_to_dPE_k, dS_to_dPE_k dS_to_dPE_k, dT_to_dPEa dT_to_dPEa, dS_to_dPEa dS_to_dPEa, pres_Z pres_Z, dT_to_dColHt_k dT_to_dColHt_k, dS_to_dColHt_k dS_to_dColHt_k, dT_to_dColHta dT_to_dColHta, dS_to_dColHta dS_to_dColHta, PE_chg PE_chg, dPEc_dKd dPEc_dKd, dPE_max dPE_max, dPEc_dKd_0 dPEc_dKd_0)¶ This subroutine calculates the change in potential energy and or derivatives for several changes in an interfaces’s diapycnal diffusivity times a timestep using the original form used in the first version of ePBL.
- Parameters
[in] kddt_h: The diffusivity at an interface times the time step and divided by the average of the thicknesses around the interface [H ~> m or kg m-2].[in] h_k: The thickness of the layer below the interface [H ~> m or kg m-2].[in] b_den_1: The first term in the denominator of the pivot for the tridiagonal solver, given by h_k plus a term that is a fraction (determined from the tridiagonal solver) of Kddt_h for the interface above [H ~> m or kg m-2].[in] dte_term: A diffusivity-independent term related to the temperature change in the layer below the interface [degC H ~> degC m or degC kg m-2].[in] dse_term: A diffusivity-independent term related to the salinity change in the layer below the interface [ppt H ~> ppt m or ppt kg m-2].[in] dt_km1_t2: A diffusivity-independent term related to the temperature change in the layer above the interface [degC].[in] ds_km1_t2: A diffusivity-independent term related to the salinity change in the layer above the interface [ppt].[in] pres_z: The hydrostatic interface pressure, which is used to relate the changes in column thickness to the energy that is radiated as gravity waves and unavailable to drive mixing [J m-2 Z-1 ~> J m-3].[in] dt_to_dpe_k: A factor (pres_lay*mass_lay*dSpec_vol/dT) relating a layer’s temperature change to the change in column potential energy, including all implicit diffusive changes in the temperatures of all the layers below [J m-2 degC-1].[in] ds_to_dpe_k: A factor (pres_lay*mass_lay*dSpec_vol/dS) relating a layer’s salinity change to the change in column potential energy, including all implicit diffusive changes in the salinities of all the layers below [J m-2 ppt-1].[in] dt_to_dpea: A factor (pres_lay*mass_lay*dSpec_vol/dT) relating a layer’s temperature change to the change in column potential energy, including all implicit diffusive changes in the temperatures of all the layers above [J m-2 degC-1].[in] ds_to_dpea: A factor (pres_lay*mass_lay*dSpec_vol/dS) relating a layer’s salinity change to the change in column potential energy, including all implicit diffusive changes in the salinities of all the layers above [J m-2 ppt-1].[in] dt_to_dcolht_k: A factor (mass_lay*dSColHtc_vol/dT) relating a layer’s temperature change to the change in column height, including all implicit diffusive changes in the temperatures of all the layers below [Z degC-1 ~> m degC-1].[in] ds_to_dcolht_k: A factor (mass_lay*dSColHtc_vol/dS) relating a layer’s salinity change to the change in column height, including all implicit diffusive changes in the salinities of all the layers below [Z ppt-1 ~> m ppt-1].[in] dt_to_dcolhta: A factor (mass_lay*dSColHtc_vol/dT) relating a layer’s temperature change to the change in column height, including all implicit diffusive changes in the temperatures of all the layers above [Z degC-1 ~> m degC-1].[in] ds_to_dcolhta: A factor (mass_lay*dSColHtc_vol/dS) relating a layer’s salinity change to the change in column height, including all implicit diffusive changes in the salinities of all the layers above [Z ppt-1 ~> m ppt-1].[out] pe_chg: The change in column potential energy from applying Kddt_h at the present interface [J m-2].[out] dpec_dkd: The partial derivative of PE_chg with Kddt_h, [J m-2 H-1 ~> J m-3 or J kg-1].[out] dpe_max: The maximum change in column potential energy that could be realized by applying a huge value of Kddt_h at the present interface [J m-2].[out] dpec_dkd_0: The partial derivative of PE_chg with Kddt_h in the limit where Kddt_h = 0 [J m-2 H-1 ~> J m-3 or J kg-1].
-
subroutine, public mom_diapyc_energy_req::diapyc_energy_req_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS) Initialize parameters and allocate memory associated with the diapycnal energy requirement module.
- Parameters
[in] time: model time[in] g: model grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: file to parse for parameter values[inout] diag: structure to regulate diagnostic outputcs: module control structure
-
subroutine, public mom_diapyc_energy_req::diapyc_energy_req_end(CS CS) Clean up and deallocate memory associated with the diapycnal energy requirement module.
- Parameters
cs: Diapycnal energy requirement control structure that will be deallocated in this subroutine.
-
-
namespace
mom_document¶ The subroutines here provide hooks for document generation functions at various levels of granularity.
Functions
-
subroutine
doc_param_none(doc doc, varname varname, desc desc, units units)¶ This subroutine handles parameter documentation with no value.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] varname: The name of the parameter being documented[in] desc: A description of the parameter being documented[in] units: The units of the parameter being documented
-
subroutine mom_document::doc_param_logical(doc doc, varname varname, desc desc, units units, val val, default default, layoutParam layoutParam, debuggingParam debuggingParam) This subroutine handles parameter documentation for logicals.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] varname: The name of the parameter being documented[in] desc: A description of the parameter being documented[in] units: The units of the parameter being documented[in] val: The value of this parameter[in] default: The default value of this parameter[in] layoutparam: If present and true, this is a layout parameter.[in] debuggingparam: If present and true, this is a debugging parameter.
-
subroutine mom_document::doc_param_logical_array(doc doc, varname varname, desc desc, units units, vals vals, default default, layoutParam layoutParam, debuggingParam debuggingParam) This subroutine handles parameter documentation for arrays of logicals.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] varname: The name of the parameter being documented[in] desc: A description of the parameter being documented[in] units: The units of the parameter being documented[in] vals: The array of values to record[in] default: The default value of this parameter[in] layoutparam: If present and true, this is a layout parameter.[in] debuggingparam: If present and true, this is a debugging parameter.
-
subroutine mom_document::doc_param_int(doc doc, varname varname, desc desc, units units, val val, default default, layoutParam layoutParam, debuggingParam debuggingParam) This subroutine handles parameter documentation for integers.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] varname: The name of the parameter being documented[in] desc: A description of the parameter being documented[in] units: The units of the parameter being documented[in] val: The value of this parameter[in] default: The default value of this parameter[in] layoutparam: If present and true, this is a layout parameter.[in] debuggingparam: If present and true, this is a debugging parameter.
-
subroutine mom_document::doc_param_int_array(doc doc, varname varname, desc desc, units units, vals vals, default default, layoutParam layoutParam, debuggingParam debuggingParam) This subroutine handles parameter documentation for arrays of integers.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] varname: The name of the parameter being documented[in] desc: A description of the parameter being documented[in] units: The units of the parameter being documented[in] vals: The array of values to record[in] default: The default value of this parameter[in] layoutparam: If present and true, this is a layout parameter.[in] debuggingparam: If present and true, this is a debugging parameter.
-
subroutine mom_document::doc_param_real(doc doc, varname varname, desc desc, units units, val val, default default, debuggingParam debuggingParam) This subroutine handles parameter documentation for reals.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] varname: The name of the parameter being documented[in] desc: A description of the parameter being documented[in] units: The units of the parameter being documented[in] val: The value of this parameter[in] default: The default value of this parameter[in] debuggingparam: If present and true, this is a debugging parameter.
-
subroutine mom_document::doc_param_real_array(doc doc, varname varname, desc desc, units units, vals vals, default default, debuggingParam debuggingParam) This subroutine handles parameter documentation for arrays of reals.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] varname: The name of the parameter being documented[in] desc: A description of the parameter being documented[in] units: The units of the parameter being documented[in] vals: The array of values to record[in] default: The default value of this parameter[in] debuggingparam: If present and true, this is a debugging parameter.
-
subroutine mom_document::doc_param_char(doc doc, varname varname, desc desc, units units, val val, default default, layoutParam layoutParam, debuggingParam debuggingParam) This subroutine handles parameter documentation for character strings.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] varname: The name of the parameter being documented[in] desc: A description of the parameter being documented[in] units: The units of the parameter being documented[in] val: The value of the parameter[in] default: The default value of this parameter[in] layoutparam: If present and true, this is a layout parameter.[in] debuggingparam: If present and true, this is a debugging parameter.
-
subroutine, public mom_document::doc_openblock(doc doc, blockName blockName, desc desc) This subroutine handles documentation for opening a parameter block.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] blockname: The name of the parameter block being opened[in] desc: A description of the parameter block being opened
-
subroutine, public mom_document::doc_closeblock(doc doc, blockName blockName) This subroutine handles documentation for closing a parameter block.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] blockname: The name of the parameter block being closed
-
subroutine mom_document::doc_param_time(doc doc, varname varname, desc desc, units units, val val, default default, layoutParam layoutParam, debuggingParam debuggingParam) This subroutine handles parameter documentation for time-type variables.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] varname: The name of the parameter being documented[in] desc: A description of the parameter being documented[in] units: The units of the parameter being documented[in] val: The value of the parameter[in] default: The default value of this parameter[in] layoutparam: If present and true, this is a layout parameter.[in] debuggingparam: If present and true, this is a debugging parameter.
-
subroutine
writemessageanddesc(doc doc, vmesg vmesg, desc desc, valueWasDefault valueWasDefault, indent indent, layoutParam layoutParam, debuggingParam debuggingParam)¶ This subroutine writes out the message and description to the documetation files.
- Parameters
[in] doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] vmesg: A message with the parameter name, units, and default value.[in] desc: A description of the parameter being documented[in] valuewasdefault: If true, this parameter has its default value[in] indent: An amount by which to indent this message[in] layoutparam: If present and true, this is a layout parameter.[in] debuggingparam: If present and true, this is a debugging parameter.
-
character(len=32) function mom_document::real_string(val val) This function returns a string with a real formatted like ‘(G)’.
- Parameters
[in] val: The value being written into a string
-
character(len=1320) function mom_document::real_array_string(vals vals, sep sep) Returns a character string of a comma-separated, compact formatted, reals e.g. “1., 2., 5*3., 5.E2”, that give the list of values.
- Return
The output string listing vals
- Parameters
[in] vals: The array of values to record[in] sep: The separator between successive values,
-
logical function mom_document::testformattedfloatisreal(str str, val val) This function tests whether a real value is encoded in a string.
- Parameters
[in] str: The string that match val[in] val: The value being tested
-
character(len=24) function mom_document::int_string(val val) This function returns a string with an integer formatted like ‘(I)’.
- Parameters
[in] val: The value being written into a string
-
character(len=24) function mom_document::logical_string(val val) This function returns a string with an logical formatted like ‘(L)’.
- Parameters
[in] val: The value being written into a string
-
character(len=mlen) function mom_document::define_string(doc doc, varName varName, valString valString, units units) This function returns a string for formatted parameter assignment.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] varname: The name of the parameter being documented[in] valstring: A string containing the value of the parameter[in] units: The units of the parameter being documented
-
character(len=mlen) function mom_document::undef_string(doc doc, varName varName, units units) This function returns a string for formatted false logicals.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] varname: The name of the parameter being documented[in] units: The units of the parameter being documented
-
subroutine, public mom_document::doc_module(doc doc, modname modname, desc desc) This subroutine handles the module documentation.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] modname: The name of the module being documented[in] desc: A description of the module being documented
-
subroutine, public mom_document::doc_subroutine(doc doc, modname modname, subname subname, desc desc) This subroutine handles the subroutine documentation.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] modname: The name of the module being documented[in] subname: The name of the subroutine being documented[in] desc: A description of the subroutine being documented
-
subroutine, public mom_document::doc_function(doc doc, modname modname, fnname fnname, desc desc) This subroutine handles the function documentation.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] modname: The name of the module being documented[in] fnname: The name of the function being documented[in] desc: A description of the function being documented
-
subroutine, public mom_document::doc_init(docFileBase docFileBase, doc doc, minimal minimal, complete complete, layout layout, debugging debugging) Initialize the parameter documentation.
- Parameters
[in] docfilebase: The base file name for this set of parameters, for example MOM_parameter_docdoc: A pointer to a structure that controls where the documentation occurs and its formatting[in] minimal: If present and true, write out the files (.short) documenting those parameters that do not take on their default values.[in] complete: If present and true, write out the (.all) files documenting all parameters[in] layout: If present and true, write out the (.layout) files documenting the layout parameters[in] debugging: If present and true, write out the (.debugging) files documenting the debugging parameters
-
subroutine
open_doc_file(doc doc)¶ This subroutine allocates and populates a structure that controls where the documentation occurs and its formatting, and opens up the files controlled by this structure.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting
-
integer function mom_document::find_unused_unit_number() Find an unused unit number, returning >0 if found, and triggering a FATAL error if not.
-
subroutine, public mom_document::doc_end(doc doc) This subroutine closes the the files controlled by doc, and sets flags in doc to indicate that parameterization is no longer permitted.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting
-
logical function mom_document::mesghasbeendocumented(doc doc, varName varName, mesg mesg) Returns true if documentation has already been written.
- Parameters
doc: A pointer to a structure that controls where the documentation occurs and its formatting[in] varname: The name of the parameter being documented[in] mesg: A message with parameter values, defaults, and descriptions to compare with the message that was written previously
Variables
-
integer, parameter mom_document::mlen = 1240 Length of interface/message strings.
-
character(len=4), parameter mom_document::string_true = 'True' A string for true logicals.
-
character(len=5), parameter mom_document::string_false = 'False' A string for false logicals.
-
subroutine
-
namespace
mom_domains¶ Describes the decomposed MOM domain and has routines for communications across PEs.
Functions
-
subroutine
pass_var_3d(array array, MOM_dom MOM_dom, sideflag sideflag, complete complete, position position, halo halo, clock clock)¶ pass_var_3d does a halo update for a three-dimensional array.
- Parameters
[inout] array: The array which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] sideflag: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH. For example, TO_EAST sends the data to the processor to the east, sothe halos on the western side are filled. TO_ALL is the default if sideflag is omitted.[in] complete: An optional argument indicating whether the halo updates should be completed before progress resumes. Omitting complete is the same as setting complete to .true.[in] position: An optional argument indicating the position. This is usally CORNER, but is CENTER by default.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
pass_var_2d(array array, MOM_dom MOM_dom, sideflag sideflag, complete complete, position position, halo halo, inner_halo inner_halo, clock clock)¶ pass_var_2d does a halo update for a two-dimensional array.
- Parameters
[inout] array: The array which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] sideflag: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if sideflag is omitted.[in] complete: An optional argument indicating whether the halo updates should be completed before progress resumes. Omitting complete is the same as setting complete to .true.[in] position: An optional argument indicating the position. This is usally CORNER, but is CENTER by default.[in] halo: The size of the halo to update - the full halo by default.[in] inner_halo: The size of an inner halo to avoid updating, or 0 to avoid updating symmetric memory computational domain points. Setting this >=0 also enforces that complete=.true.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
integer function mom_domains::pass_var_start_2d(array array, MOM_dom MOM_dom, sideflag sideflag, position position, complete complete, halo halo, clock clock) pass_var_start_2d starts a halo update for a two-dimensional array.
- Return
The integer index for this update.
- Parameters
[inout] array: The array which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] sideflag: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if sideflag is omitted.[in] position: An optional argument indicating the position. This is usally CORNER, but is CENTER by default.[in] complete: An optional argument indicating whether the halo updates should be completed before progress resumes. Omitting complete is the same as setting complete to .true.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
integer function mom_domains::pass_var_start_3d(array array, MOM_dom MOM_dom, sideflag sideflag, position position, complete complete, halo halo, clock clock) pass_var_start_3d starts a halo update for a three-dimensional array.
- Return
The integer index for this update.
- Parameters
[inout] array: The array which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] sideflag: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if sideflag is omitted.[in] position: An optional argument indicating the position. This is usally CORNER, but is CENTER by default.[in] complete: An optional argument indicating whether the halo updates should be completed before progress resumes. Omitting complete is the same as setting complete to .true.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
pass_var_complete_2d(id_update id_update, array array, MOM_dom MOM_dom, sideflag sideflag, position position, halo halo, clock clock)¶ pass_var_complete_2d completes a halo update for a two-dimensional array.
- Parameters
[in] id_update: The integer id of this update which has been returned from a previous call to pass_var_start.[inout] array: The array which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] sideflag: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if sideflag is omitted.[in] position: An optional argument indicating the position. This is usally CORNER, but is CENTER by default.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
pass_var_complete_3d(id_update id_update, array array, MOM_dom MOM_dom, sideflag sideflag, position position, halo halo, clock clock)¶ pass_var_complete_3d completes a halo update for a three-dimensional array.
- Parameters
[in] id_update: The integer id of this update which has been returned from a previous call to pass_var_start.[inout] array: The array which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] sideflag: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if sideflag is omitted.[in] position: An optional argument indicating the position. This is usally CORNER, but is CENTER by default.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
pass_vector_2d(u_cmpt u_cmpt, v_cmpt v_cmpt, MOM_dom MOM_dom, direction direction, stagger stagger, complete complete, halo halo, clock clock)¶ pass_vector_2d does a halo update for a pair of two-dimensional arrays representing the compontents of a two-dimensional horizontal vector.
- Parameters
[inout] u_cmpt: The nominal zonal (u) component of the vector pair which is having its halos points exchanged.[inout] v_cmpt: The nominal meridional (v) component of the vector pair which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] direction: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH, possibly plus SCALAR_PAIR if these are paired non-directional scalars discretized at the typical vector component locations. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if omitted.[in] stagger: An optional flag, which may be one of A_GRID, BGRID_NE, or CGRID_NE, indicating where the two components of the vector are discretized. Omitting stagger is the same as setting it to CGRID_NE.[in] complete: An optional argument indicating whether the halo updates should be completed before progress resumes. Omitting complete is the same as setting complete to .true.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
fill_vector_symmetric_edges_2d(u_cmpt u_cmpt, v_cmpt v_cmpt, MOM_dom MOM_dom, stagger stagger, scalar scalar, clock clock)¶ fill_vector_symmetric_edges_2d does an usual set of halo updates that only fill in the values at the edge of a pair of symmetric memory two-dimensional arrays representing the compontents of a two-dimensional horizontal vector. If symmetric memory is not being used, this subroutine does nothing except to possibly turn optional cpu clocks on or off.
- Parameters
[inout] u_cmpt: The nominal zonal (u) component of the vector pair which is having its halos points exchanged.[inout] v_cmpt: The nominal meridional (v) component of the vector pair which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] stagger: An optional flag, which may be one of A_GRID, BGRID_NE, or CGRID_NE, indicating where the two components of the vector are discretized. Omitting stagger is the same as setting it to CGRID_NE.[in] scalar: An optional argument indicating whether.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
pass_vector_3d(u_cmpt u_cmpt, v_cmpt v_cmpt, MOM_dom MOM_dom, direction direction, stagger stagger, complete complete, halo halo, clock clock)¶ pass_vector_3d does a halo update for a pair of three-dimensional arrays representing the compontents of a three-dimensional horizontal vector.
- Parameters
[inout] u_cmpt: The nominal zonal (u) component of the vector pair which is having its halos points exchanged.[inout] v_cmpt: The nominal meridional (v) component of the vector pair which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] direction: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH, possibly plus SCALAR_PAIR if these are paired non-directional scalars discretized at the typical vector component locations. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if omitted.[in] stagger: An optional flag, which may be one of A_GRID, BGRID_NE, or CGRID_NE, indicating where the two components of the vector are discretized. Omitting stagger is the same as setting it to CGRID_NE.[in] complete: An optional argument indicating whether the halo updates should be completed before progress resumes. Omitting complete is the same as setting complete to .true.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
integer function mom_domains::pass_vector_start_2d(u_cmpt u_cmpt, v_cmpt v_cmpt, MOM_dom MOM_dom, direction direction, stagger stagger, complete complete, halo halo, clock clock) pass_vector_start_2d starts a halo update for a pair of two-dimensional arrays representing the compontents of a two-dimensional horizontal vector.
- Return
The integer index for this update.
- Parameters
[inout] u_cmpt: The nominal zonal (u) component of the vector pair which is having its halos points exchanged.[inout] v_cmpt: The nominal meridional (v) component of the vector pair which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] direction: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH, possibly plus SCALAR_PAIR if these are paired non-directional scalars discretized at the typical vector component locations. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if omitted.[in] stagger: An optional flag, which may be one of A_GRID, BGRID_NE, or CGRID_NE, indicating where the two components of the vector are discretized. Omitting stagger is the same as setting it to CGRID_NE.[in] complete: An optional argument indicating whether the halo updates should be completed before progress resumes. Omitting complete is the same as setting complete to .true.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
integer function mom_domains::pass_vector_start_3d(u_cmpt u_cmpt, v_cmpt v_cmpt, MOM_dom MOM_dom, direction direction, stagger stagger, complete complete, halo halo, clock clock) pass_vector_start_3d starts a halo update for a pair of three-dimensional arrays representing the compontents of a three-dimensional horizontal vector.
- Return
The integer index for this update.
- Parameters
[inout] u_cmpt: The nominal zonal (u) component of the vector pair which is having its halos points exchanged.[inout] v_cmpt: The nominal meridional (v) component of the vector pair which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] direction: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH, possibly plus SCALAR_PAIR if these are paired non-directional scalars discretized at the typical vector component locations. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if omitted.[in] stagger: An optional flag, which may be one of A_GRID, BGRID_NE, or CGRID_NE, indicating where the two components of the vector are discretized. Omitting stagger is the same as setting it to CGRID_NE.[in] complete: An optional argument indicating whether the halo updates should be completed before progress resumes. Omitting complete is the same as setting complete to .true.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
pass_vector_complete_2d(id_update id_update, u_cmpt u_cmpt, v_cmpt v_cmpt, MOM_dom MOM_dom, direction direction, stagger stagger, halo halo, clock clock)¶ pass_vector_complete_2d completes a halo update for a pair of two-dimensional arrays representing the compontents of a two-dimensional horizontal vector.
- Parameters
[in] id_update: The integer id of this update which has been returned from a previous call to pass_var_start.[inout] u_cmpt: The nominal zonal (u) component of the vector pair which is having its halos points exchanged.[inout] v_cmpt: The nominal meridional (v) component of the vector pair which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] direction: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH, possibly plus SCALAR_PAIR if these are paired non-directional scalars discretized at the typical vector component locations. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if omitted.[in] stagger: An optional flag, which may be one of A_GRID, BGRID_NE, or CGRID_NE, indicating where the two components of the vector are discretized. Omitting stagger is the same as setting it to CGRID_NE.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
pass_vector_complete_3d(id_update id_update, u_cmpt u_cmpt, v_cmpt v_cmpt, MOM_dom MOM_dom, direction direction, stagger stagger, halo halo, clock clock)¶ pass_vector_complete_3d completes a halo update for a pair of three-dimensional arrays representing the compontents of a three-dimensional horizontal vector.
- Parameters
[in] id_update: The integer id of this update which has been returned from a previous call to pass_var_start.[inout] u_cmpt: The nominal zonal (u) component of the vector pair which is having its halos points exchanged.[inout] v_cmpt: The nominal meridional (v) component of the vector pair which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] direction: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH, possibly plus SCALAR_PAIR if these are paired non-directional scalars discretized at the typical vector component locations. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if omitted.[in] stagger: An optional flag, which may be one of A_GRID, BGRID_NE, or CGRID_NE, indicating where the two components of the vector are discretized. Omitting stagger is the same as setting it to CGRID_NE.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
create_var_group_pass_2d(group group, array array, MOM_dom MOM_dom, sideflag sideflag, position position, halo halo, clock clock)¶ create_var_group_pass_2d sets up a group of two-dimensional array halo updates.
- Parameters
[inout] group: The data type that store information for group update. This data will be used in do_group_pass.[inout] array: The array which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] sideflag: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if sideflag is omitted.[in] position: An optional argument indicating the position. This is usally CORNER, but is CENTER by default.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
create_var_group_pass_3d(group group, array array, MOM_dom MOM_dom, sideflag sideflag, position position, halo halo, clock clock)¶ create_var_group_pass_3d sets up a group of three-dimensional array halo updates.
- Parameters
[inout] group: The data type that store information for group update. This data will be used in do_group_pass.[inout] array: The array which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] sideflag: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if sideflag is omitted.[in] position: An optional argument indicating the position. This is usally CORNER, but is CENTER by default.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
create_vector_group_pass_2d(group group, u_cmpt u_cmpt, v_cmpt v_cmpt, MOM_dom MOM_dom, direction direction, stagger stagger, halo halo, clock clock)¶ create_vector_group_pass_2d sets up a group of two-dimensional vector halo updates.
- Parameters
[inout] group: The data type that store information for group update. This data will be used in do_group_pass.[inout] u_cmpt: The nominal zonal (u) component of the vector pair which is having its halos points exchanged.[inout] v_cmpt: The nominal meridional (v) component of the vector pair which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent[in] direction: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH, possibly plus SCALAR_PAIR if these are paired non-directional scalars discretized at the typical vector component locations. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if omitted.[in] stagger: An optional flag, which may be one of A_GRID, BGRID_NE, or CGRID_NE, indicating where the two components of the vector are discretized. Omitting stagger is the same as setting it to CGRID_NE.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine
create_vector_group_pass_3d(group group, u_cmpt u_cmpt, v_cmpt v_cmpt, MOM_dom MOM_dom, direction direction, stagger stagger, halo halo, clock clock)¶ create_vector_group_pass_3d sets up a group of three-dimensional vector halo updates.
- Parameters
[inout] group: The data type that store information for group update. This data will be used in do_group_pass.[inout] u_cmpt: The nominal zonal (u) component of the vector pair which is having its halos points exchanged.[inout] v_cmpt: The nominal meridional (v) component of the vector pair which is having its halos points exchanged.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] direction: An optional integer indicating which directions the data should be sent. It is TO_ALL or the sum of any of TO_EAST, TO_WEST, TO_NORTH, and TO_SOUTH, possibly plus SCALAR_PAIR if these are paired non-directional scalars discretized at the typical vector component locations. For example, TO_EAST sends the data to the processor to the east, so the halos on the western side are filled. TO_ALL is the default if omitted.[in] stagger: An optional flag, which may be one of A_GRID, BGRID_NE, or CGRID_NE, indicating where the two components of the vector are discretized. Omitting stagger is the same as setting it to CGRID_NE.[in] halo: The size of the halo to update - the full halo by default.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine, public mom_domains::do_group_pass(group group, MOM_dom MOM_dom, clock clock) do_group_pass carries out a group halo update.
- Parameters
[inout] group: The data type that store information for group update. This data will be used in do_group_pass.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine, public mom_domains::start_group_pass(group group, MOM_dom MOM_dom, clock clock) start_group_pass starts out a group halo update.
- Parameters
[inout] group: The data type that store information for group update. This data will be used in do_group_pass.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine, public mom_domains::complete_group_pass(group group, MOM_dom MOM_dom, clock clock) complete_group_pass completes a group halo update.
- Parameters
[inout] group: The data type that store information for group update. This data will be used in do_group_pass.[inout] mom_dom: The MOM_domain_type containing the mpp_domain needed to determine where data should be sent.[in] clock: The handle for a cpu time clock that should be started then stopped to time this routine.
-
subroutine, public mom_domains::mom_domains_init(MOM_dom MOM_dom, param_file param_file, symmetric symmetric, static_memory static_memory, NIHALO NIHALO, NJHALO NJHALO, NIGLOBAL NIGLOBAL, NJGLOBAL NJGLOBAL, NIPROC NIPROC, NJPROC NJPROC, min_halo min_halo, domain_name domain_name, include_name include_name, param_suffix param_suffix) MOM_domains_init initalizes a MOM_domain_type variable, based on the information read in from a param_file_type, and optionally returns data describing various’ properties of the domain type.
- Parameters
mom_dom: A pointer to the MOM_domain_type being defined here.[in] param_file: A structure to parse for run-time parameters[in] symmetric: If present, this specifies whether this domain is symmetric, regardless of whether the macro SYMMETRIC_MEMORY_ is defined.[in] static_memory: If present and true, this domain type is set up for static memory and error checking of various input values is performed against those in the input file.[in] nihalo: Default halo sizes, required with static memory.[in] njhalo: Default halo sizes, required with static memory.[in] niglobal: Total domain sizes, required with static memory.[in] njglobal: Total domain sizes, required with static memory.[in] niproc: Processor counts, required with static memory.[in] njproc: Processor counts, required with static memory.[inout] min_halo: If present, this sets the minimum halo size for this domain in the i- and j- directions, and returns the actual halo size used.[in] domain_name: A name for this domain, “MOM” if missing.[in] include_name: A name for model’s include file, “MOM_memory.h” if missing.[in] param_suffix: A suffix to apply to layout-specific parameters.
-
subroutine
clone_md_to_md(MD_in MD_in, MOM_dom MOM_dom, min_halo min_halo, halo_size halo_size, symmetric symmetric, domain_name domain_name)¶ clone_MD_to_MD copies one MOM_domain_type into another, while allowing some properties of the new type to differ from the original one.
- Parameters
[in] md_in: An existing MOM_domainmom_dom: A pointer to a MOM_domain that will be allocated if it is unassociated, and will have data copied from MD_in[inout] min_halo: If present, this sets the[in] halo_size: If present, this sets the halo size for the domian in the i- and j-directions. min_halo and halo_size can not both be present.[in] symmetric: If present, this specifies whether the new domain is symmetric, regardless of whether the macro SYMMETRIC_MEMORY_ is defined.[in] domain_name: A name for the new domain, “MOM”
-
subroutine
clone_md_to_d2d(MD_in MD_in, mpp_domain mpp_domain, min_halo min_halo, halo_size halo_size, symmetric symmetric, domain_name domain_name)¶ clone_MD_to_d2D uses information from a MOM_domain_type to create a new domain2d type, while allowing some properties of the new type to differ from the original one.
- Parameters
[in] md_in: An existing MOM_domain to be cloned[inout] mpp_domain: The new mpp_domain to be set up[inout] min_halo: If present, this sets the[in] halo_size: If present, this sets the halo size for the domian in the i- and j-directions. min_halo and halo_size can not both be present.[in] symmetric: If present, this specifies whether the new domain is symmetric, regardless of whether the macro SYMMETRIC_MEMORY_ is defined.[in] domain_name: A name for the new domain, “MOM”
-
subroutine, public mom_domains::get_domain_extent(Domain Domain, isc isc, iec iec, jsc jsc, jec jec, isd isd, ied ied, jsd jsd, jed jed, isg isg, ieg ieg, jsg jsg, jeg jeg, idg_offset idg_offset, jdg_offset jdg_offset, symmetric symmetric, local_indexing local_indexing, index_offset index_offset) Returns various data that has been stored in a MOM_domain_type.
- Parameters
[in] domain: The MOM domain from which to extract information[out] isc: The start i-index of the computational domain[out] iec: The end i-index of the computational domain[out] jsc: The start j-index of the computational domain[out] jec: The end j-index of the computational domain[out] isd: The start i-index of the data domain[out] ied: The end i-index of the data domain[out] jsd: The start j-index of the data domain[out] jed: The end j-index of the data domain[out] isg: The start i-index of the global domain[out] ieg: The end i-index of the global domain[out] jsg: The start j-index of the global domain[out] jeg: The end j-index of the global domain[out] idg_offset: The offset between the corresponding global and data i-index spaces.[out] jdg_offset: The offset between the corresponding global and data j-index spaces.[out] symmetric: True if symmetric memory is used.[in] local_indexing: If true, local tracer array indices start at 1, as in most MOM6 code.[in] index_offset: A fixed additional offset to all indices. This can be useful for some types of debugging with dynamic memory allocation.
-
subroutine, public mom_domains::get_domain_extent_dsamp2(Domain Domain, isc_d2 isc_d2, iec_d2 iec_d2, jsc_d2 jsc_d2, jec_d2 jec_d2, isd_d2 isd_d2, ied_d2 ied_d2, jsd_d2 jsd_d2, jed_d2 jed_d2, isg_d2 isg_d2, ieg_d2 ieg_d2, jsg_d2 jsg_d2, jeg_d2 jeg_d2) - Parameters
[in] domain: The MOM domain from which to extract information[out] isc_d2: The start i-index of the computational domain[out] iec_d2: The end i-index of the computational domain[out] jsc_d2: The start j-index of the computational domain[out] jec_d2: The end j-index of the computational domain[out] isd_d2: The start i-index of the data domain[out] ied_d2: The end i-index of the data domain[out] jsd_d2: The start j-index of the data domain[out] jed_d2: The end j-index of the data domain[out] isg_d2: The start i-index of the global domain[out] ieg_d2: The end i-index of the global domain[out] jsg_d2: The start j-index of the global domain[out] jeg_d2: The end j-index of the global domain
-
subroutine, public mom_domains::get_simple_array_i_ind(domain domain, size size, is is, ie ie, symmetric symmetric) Return the (potentially symmetric) computational domain i-bounds for an array passed without index specifications (i.e. indices start at 1) based on an array size.
- Parameters
[in] domain: MOM domain from which to extract information[in] size: The i-array size[out] is: The computational domain starting i-index.[out] ie: The computational domain ending i-index.[in] symmetric: If present, indicates whether symmetric sizes can be considered.
-
subroutine, public mom_domains::get_simple_array_j_ind(domain domain, size size, js js, je je, symmetric symmetric) Return the (potentially symmetric) computational domain j-bounds for an array passed without index specifications (i.e. indices start at 1) based on an array size.
- Parameters
[in] domain: MOM domain from which to extract information[in] size: The j-array size[out] js: The computational domain starting j-index.[out] je: The computational domain ending j-index.[in] symmetric: If present, indicates whether symmetric sizes can be considered.
-
subroutine, public mom_domains::get_global_shape(domain domain, niglobal niglobal, njglobal njglobal) Returns the global shape of h-point arrays.
- Parameters
[in] domain: MOM domain[out] niglobal: i-index global size of h-point arrays[out] njglobal: j-index global size of h-point arrays
Variables
-
integer, parameter, public mom_domains::to_all = To_East + To_West + To_North + To_South A flag for passing in all directions.
-
subroutine
-
namespace
mom_dyn_horgrid¶ Contains a shareable dynamic type for describing horizontal grids and metric data and utilty routines that work on this type.
Functions
-
subroutine, public mom_dyn_horgrid::create_dyn_horgrid(G G, HI HI, bathymetry_at_vel bathymetry_at_vel) Allocate memory used by the dyn_horgrid_type and related structures.
- Parameters
g: A pointer to the dynamic horizontal grid type[in] hi: A hor_index_type for array extents[in] bathymetry_at_vel: If true, there are separate values for the basin depths at velocity points. Otherwise the effects of topography are entirely determined from thickness points.
-
subroutine, public mom_dyn_horgrid::rescale_dyn_horgrid_bathymetry(G G, m_in_new_units m_in_new_units) rescale_dyn_horgrid_bathymetry permits a change in the internal units for the bathymetry on the grid, both rescaling the depths and recording the new internal depth units.
- Parameters
[inout] g: The dynamic horizontal grid type[in] m_in_new_units: The new internal representation of 1 m depth.
-
subroutine, public mom_dyn_horgrid::set_derived_dyn_horgrid(G G) set_derived_dyn_horgrid calculates metric terms that are derived from other metrics.
- Parameters
[inout] g: The dynamic horizontal grid type
-
real function mom_dyn_horgrid::adcroft_reciprocal(val val) Adcroft_reciprocal(x) = 1/x for |x|>0 or 0 for x=0.
- Return
The Adcroft reciprocal of val.
- Parameters
[in] val: The value being inverted.
-
subroutine, public mom_dyn_horgrid::destroy_dyn_horgrid(G G) Release memory used by the dyn_horgrid_type and related structures.
- Parameters
g: The dynamic horizontal grid type
-
-
namespace
mom_dynamics_split_rk2¶ Time step the adiabatic dynamic core of MOM using RK2 method.
This file time steps the adiabatic dynamic core by splitting between baroclinic and barotropic modes. It uses a pseudo-second order Runge-Kutta time stepping scheme for the baroclinic momentum equation and a forward-backward coupling between the baroclinic momentum and continuity equations. This split time-stepping scheme is described in detail in Hallberg (JCP, 1997). Additional issues related to exact tracer conservation and how to ensure consistency between the barotropic and layered estimates of the free surface height are described in Hallberg and Adcroft (Ocean Modelling, 2009). This was the time stepping code that is used for most GOLD applications, including GFDL’s ESM2G Earth system model, and all of the examples provided with the MOM code (although several of these solutions are routinely verified by comparison with the slower unsplit schemes).
The subroutine step_MOM_dyn_split_RK2 actually does the time stepping, while register_restarts_dyn_split_RK2 sets the fields that are found in a full restart file with this scheme, and initialize_dyn_split_RK2 initializes the cpu clocks that are used in this module. For largely historical reasons, this module does not have its own control structure, but shares the same control structure with MOM.F90 and the other MOM_dynamics_… modules.
Unnamed Group
-
integer
id_clock_cor¶ CPU time clock IDs.
-
integer
id_clock_pres¶ CPU time clock IDs.
-
integer
id_clock_vertvisc¶ CPU time clock IDs.
-
integer
id_clock_horvisc¶ CPU time clock IDs.
-
integer
id_clock_mom_update¶ CPU time clock IDs.
-
integer
id_clock_continuity¶ CPU time clock IDs.
-
integer
id_clock_thick_diff¶ CPU time clock IDs.
-
integer
id_clock_btstep¶ CPU time clock IDs.
-
integer
id_clock_btcalc¶ CPU time clock IDs.
-
integer
id_clock_btforce¶ CPU time clock IDs.
-
integer
id_clock_pass¶ CPU time clock IDs.
-
integer
id_clock_pass_init¶ CPU time clock IDs.
-
subroutine, public mom_dynamics_split_rk2::step_mom_dyn_split_rk2(u u, v v, h h, tv tv, visc visc, Time_local Time_local, dt dt, forces forces, p_surf_begin p_surf_begin, p_surf_end p_surf_end, uh uh, vh vh, uhtr uhtr, vhtr vhtr, eta_av eta_av, G G, GV GV, US US, CS CS, calc_dtbt calc_dtbt, VarMix VarMix, MEKE MEKE, thickness_diffuse_CSp thickness_diffuse_CSp, Waves Waves) RK2 splitting for time stepping MOM adiabatic dynamics.
- Parameters
[inout] g: ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] u: zonal velocity [m s-1][inout] v: merid velocity [m s-1][inout] h: layer thickness [H ~> m or kg m-2][in] tv: thermodynamic type[inout] visc: vertical visc, bottom drag, and related[in] time_local: model time at end of time step[in] dt: time step [s][in] forces: A structure with the driving mechanical forcesp_surf_begin: surf pressure at start of this dynamic time step [Pa]p_surf_end: surf pressure at end of this dynamic time step [Pa][inout] uh: zonal volume/mass transport[inout] vh: merid volume/mass transport[inout] uhtr: accumulatated zonal volume/mass transport[inout] vhtr: accumulatated merid volume/mass transport[out] eta_av: free surface height or column mass time averaged over time step [H ~> m or kg m-2]cs: module control structure[in] calc_dtbt: if true, recalculate barotropic time stepvarmix: specify the spatially varying viscositiesmeke: related to mesoscale eddy kinetic energy paramthickness_diffuse_csp: Pointer to a structure containing interface height diffusivitieswaves: A pointer to a structure containing fields related to the surface wave conditions
-
subroutine, public mom_dynamics_split_rk2::register_restarts_dyn_split_rk2(HI HI, GV GV, param_file param_file, CS CS, restart_CS restart_CS, uh uh, vh vh) This subroutine sets up any auxiliary restart variables that are specific to the unsplit time stepping scheme. All variables registered here should have the ability to be recreated if they are not present in a restart file.
- Parameters
[in] hi: Horizontal index structure[in] gv: ocean vertical grid structure[in] param_file: parameter filecs: module control structurerestart_cs: restart control structure[inout] uh: zonal volume/mass transport [H m2 s-1 ~> m3 s-1 or kg s-1][inout] vh: merid volume/mass transport [H m2 s-1 ~> m3 s-1 or kg s-1]
-
subroutine, public mom_dynamics_split_rk2::initialize_dyn_split_rk2(u u, v v, h h, uh uh, vh vh, eta eta, Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS, restart_CS restart_CS, dt dt, Accel_diag Accel_diag, Cont_diag Cont_diag, MIS MIS, VarMix VarMix, MEKE MEKE, thickness_diffuse_CSp thickness_diffuse_CSp, OBC OBC, update_OBC_CSp update_OBC_CSp, ALE_CSp ALE_CSp, setVisc_CSp setVisc_CSp, visc visc, dirs dirs, ntrunc ntrunc, calc_dtbt calc_dtbt) This subroutine initializes all of the variables that are used by this dynamic core, including diagnostics and the cpu clocks.
- Parameters
[inout] g: ocean grid structure[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] u: zonal velocity [m s-1][inout] v: merid velocity [m s-1][inout] h: layer thickness [H ~> m or kg m-2][inout] uh: zonal volume/mass transport [H m2 s-1 ~> m3 s-1 or kg s-1][inout] vh: merid volume/mass transport [H m2 s-1 ~> m3 s-1 or kg s-1][inout] eta: free surface height or column mass [H ~> m or kg m-2][in] time: current model time[in] param_file: parameter file for parsing[inout] diag: to control diagnosticscs: module control structurerestart_cs: restart control structure[in] dt: time step [s][inout] accel_diag: points to momentum equation terms for budget analysis[inout] cont_diag: points to terms in continuity equation[inout] mis: “MOM6 internal state” used to pass diagnostic pointersvarmix: points to spatially variable viscositiesmeke: points to mesoscale eddy kinetic energy fieldsthickness_diffuse_csp: Pointer to the control structure used for the isopycnal height diffusive transport.obc: points to OBC related fieldsupdate_obc_csp: points to OBC update related fieldsale_csp: points to ALE control structuresetvisc_csp: points to the set_visc control structure.[inout] visc: vertical viscosities, bottom drag, and related[in] dirs: contains directory paths[inout] ntrunc: A target for the variable that records the number of times the velocity is truncated (this should be 0).[out] calc_dtbt: If true, recalculate the barotropic time step
-
subroutine, public mom_dynamics_split_rk2::end_dyn_split_rk2(CS CS) Close the dyn_split_RK2 module.
- Parameters
cs: module control structure
-
integer
-
namespace
mom_dynamics_unsplit¶ Time steps the ocean dynamics with an unsplit quasi 3rd order scheme.
Unnamed Group
-
integer
id_clock_cor¶ CPU time clock IDs.
-
integer
id_clock_pres¶ CPU time clock IDs.
-
integer
id_clock_vertvisc¶ CPU time clock IDs.
-
integer
id_clock_continuity¶ CPU time clock IDs.
-
integer
id_clock_horvisc¶ CPU time clock IDs.
-
integer
id_clock_mom_update¶ CPU time clock IDs.
-
integer
id_clock_pass¶ CPU time clock IDs.
-
integer
id_clock_pass_init¶ CPU time clock IDs.
-
subroutine, public mom_dynamics_unsplit::step_mom_dyn_unsplit(u u, v v, h h, tv tv, visc visc, Time_local Time_local, dt dt, forces forces, p_surf_begin p_surf_begin, p_surf_end p_surf_end, uh uh, vh vh, uhtr uhtr, vhtr vhtr, eta_av eta_av, G G, GV GV, US US, CS CS, VarMix VarMix, MEKE MEKE, Waves Waves) Step the MOM6 dynamics using an unsplit mixed 2nd order (for continuity) and 3rd order (for the inviscid momentum equations) order scheme.
- Parameters
[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[inout] u: The zonal velocity [m s-1].[inout] v: The meridional velocity [m s-1].[inout] h: Layer thicknesses [H ~> m or kg m-2].[in] tv: A structure pointing to various thermodynamic variables.[inout] visc: A structure containing vertical viscosities, bottom drag viscosities, and related fields.[in] time_local: The model time at the end of the time step.[in] dt: The dynamics time step [s].[in] forces: A structure with the driving mechanical forcesp_surf_begin: A pointer (perhaps NULL) to the surface pressure at the start of this dynamic step [Pa].p_surf_end: A pointer (perhaps NULL) to the surface pressure at the end of this dynamic step [Pa].[inout] uh: The zonal volume or mass transport [H m2 s-1 ~> m3 or kg s-1].[inout] vh: The meridional volume or mass transport [H m2 s-1 ~> m3 or kg s-1].[inout] uhtr: The accumulated zonal volume or mass transport since the last tracer advection [H m2 ~> m3 or kg].[inout] vhtr: The accumulated meridional volume or mass transport since the last tracer advection [H m2 ~> m3 or kg].[out] eta_av: The time-mean free surface height or column mass [H ~> m or kg m-2].cs: The control structure set up by initialize_dyn_unsplit.varmix: A pointer to a structure with fields that specify the spatially variable viscosities.meke: A pointer to a structure containing fields related to the Mesoscale Eddy Kinetic Energy.waves: A pointer to a structure containing fields related to the surface wave conditions
-
subroutine, public mom_dynamics_unsplit::register_restarts_dyn_unsplit(HI HI, GV GV, param_file param_file, CS CS, restart_CS restart_CS) Allocate the control structure for this module, allocates memory in it, and registers any auxiliary restart variables that are specific to the unsplit time stepping scheme.
All variables registered here should have the ability to be recreated if they are not present in a restart file.
- Parameters
[in] hi: A horizontal index type structure.[in] gv: The ocean’s vertical grid structure.[in] param_file: A structure to parse for run-time parameters.cs: The control structure set up by initialize_dyn_unsplit.restart_cs: A pointer to the restart control structure.
-
subroutine, public mom_dynamics_unsplit::initialize_dyn_unsplit(u u, v v, h h, Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS, restart_CS restart_CS, Accel_diag Accel_diag, Cont_diag Cont_diag, MIS MIS, MEKE MEKE, OBC OBC, update_OBC_CSp update_OBC_CSp, ALE_CSp ALE_CSp, setVisc_CSp setVisc_CSp, visc visc, dirs dirs, ntrunc ntrunc) Initialize parameters and allocate memory associated with the unsplit dynamics module.
- Parameters
[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[inout] u: The zonal velocity [m s-1].[inout] v: The meridional velocity [m s-1].[inout] h: Layer thicknesses [H ~> m or kg m-2][in] time: The current model time.[in] param_file: A structure to parse for run-time parameters.[inout] diag: A structure that is used to regulate diagnostic output.cs: The control structure set up by initialize_dyn_unsplit.restart_cs: A pointer to the restart control structure.[inout] accel_diag: A set of pointers to the various accelerations in the momentum equations, which can be used for later derived diagnostics, like energy budgets.[inout] cont_diag: A structure with pointers to various terms in the continuity equations.[inout] mis: The “MOM6 Internal State” structure, used to pass around pointers to various arrays for diagnostic purposes.meke: MEKE dataobc: If open boundary conditions are used, this points to the ocean_OBC_type that was set up in MOM_initialization.update_obc_csp: If open boundary condition updates are used, this points to the appropriate control structure.ale_csp: This points to the ALE control structure.setvisc_csp: This points to the set_visc control structure.[inout] visc: A structure containing vertical viscosities, bottom drag viscosities, and related fields.[in] dirs: A structure containing several relevant directory paths.[inout] ntrunc: A target for the variable that records the number of times the velocity is truncated (this should be 0).
-
subroutine, public mom_dynamics_unsplit::end_dyn_unsplit(CS CS) Clean up and deallocate memory associated with the unsplit dynamics module.
- Parameters
cs: unsplit dynamics control structure that will be deallocated in this subroutine.
-
integer
-
namespace
mom_dynamics_unsplit_rk2¶ Time steps the ocean dynamics with an unsplit quasi 2nd order Runge-Kutta scheme.
Unnamed Group
-
integer
id_clock_cor¶ CPU time clock IDs.
-
integer
id_clock_pres¶ CPU time clock IDs.
-
integer
id_clock_vertvisc¶ CPU time clock IDs.
-
integer
id_clock_horvisc¶ CPU time clock IDs.
-
integer
id_clock_continuity¶ CPU time clock IDs.
-
integer
id_clock_mom_update¶ CPU time clock IDs.
-
integer
id_clock_pass¶ CPU time clock IDs.
-
integer
id_clock_pass_init¶ CPU time clock IDs.
-
subroutine, public mom_dynamics_unsplit_rk2::step_mom_dyn_unsplit_rk2(u_in u_in, v_in v_in, h_in h_in, tv tv, visc visc, Time_local Time_local, dt dt, forces forces, p_surf_begin p_surf_begin, p_surf_end p_surf_end, uh uh, vh vh, uhtr uhtr, vhtr vhtr, eta_av eta_av, G G, GV GV, US US, CS CS, VarMix VarMix, MEKE MEKE) Step the MOM6 dynamics using an unsplit quasi-2nd order Runge-Kutta scheme.
- Parameters
[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[inout] u_in: The input and output zonal velocity [m s-1].[inout] v_in: The input and output meridional velocity [m s-1].[inout] h_in: The input and output layer thicknesses, [H ~> m or kg m-2], depending on whether the Boussinesq approximation is made.[in] tv: A structure pointing to various thermodynamic variables.[inout] visc: A structure containing vertical viscosities, bottom drag viscosities, and related fields.[in] time_local: The model time at the end of the time step.[in] dt: The baroclinic dynamics time step [s].[in] forces: A structure with the driving mechanical forcesp_surf_begin: A pointer (perhaps NULL) to the surface pressure at the beginning of this dynamic step [Pa].p_surf_end: A pointer (perhaps NULL) to the surface pressure at the end of this dynamic step [Pa].[inout] uh: The zonal volume or mass transport [H m2 s-1 ~> m3 or kg s-1].[inout] vh: The meridional volume or mass transport [H m2 s-1 ~> m3 or kg s-1].[inout] uhtr: The accumulated zonal volume or mass transport since the last tracer advection [H m2 ~> m3 or kg].[inout] vhtr: The accumulated meridional volume or mass transport since the last tracer advection [H m2 ~> m3 or kg].[out] eta_av: The time-mean free surface height or column mass [H ~> m or kg m-2].cs: The control structure set up by initialize_dyn_unsplit_RK2.varmix: A pointer to a structure with fields that specify the spatially variable viscosities.meke: A pointer to a structure containing fields related to the Mesoscale Eddy Kinetic Energy.
-
subroutine, public mom_dynamics_unsplit_rk2::register_restarts_dyn_unsplit_rk2(HI HI, GV GV, param_file param_file, CS CS, restart_CS restart_CS) Allocate the control structure for this module, allocates memory in it, and registers any auxiliary restart variables that are specific to the unsplit RK2 time stepping scheme.
All variables registered here should have the ability to be recreated if they are not present in a restart file.
- Parameters
[in] hi: A horizontal index type structure.[in] gv: The ocean’s vertical grid structure.[in] param_file: A structure to parse for run-time parameters.cs: The control structure set up by initialize_dyn_unsplit_RK2.restart_cs: A pointer to the restart control structure.
-
subroutine, public mom_dynamics_unsplit_rk2::initialize_dyn_unsplit_rk2(u u, v v, h h, Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS, restart_CS restart_CS, Accel_diag Accel_diag, Cont_diag Cont_diag, MIS MIS, MEKE MEKE, OBC OBC, update_OBC_CSp update_OBC_CSp, ALE_CSp ALE_CSp, setVisc_CSp setVisc_CSp, visc visc, dirs dirs, ntrunc ntrunc) Initialize parameters and allocate memory associated with the unsplit RK2 dynamics module.
- Parameters
[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[inout] u: The zonal velocity [m s-1].[inout] v: The meridional velocity [m s-1].[inout] h: Layer thicknesses [H ~> m or kg m-2][in] time: The current model time.[in] param_file: A structure to parse for run-time parameters.[inout] diag: A structure that is used to regulate diagnostic output.cs: The control structure set up by initialize_dyn_unsplit_RK2.restart_cs: A pointer to the restart control structure.[inout] accel_diag: A set of pointers to the various accelerations in the momentum equations, which can be used for later derived diagnostics, like energy budgets.[inout] cont_diag: A structure with pointers to various terms in the continuity equations.[inout] mis: The “MOM6 Internal State” structure, used to pass around pointers to various arrays for diagnostic purposes.meke: MEKE dataobc: If open boundary conditions are used, this points to the ocean_OBC_type that was set up in MOM_initialization.update_obc_csp: If open boundary condition updates are used, this points to the appropriate control structure.ale_csp: This points to the ALE control structure.setvisc_csp: This points to the set_visc control structure.[inout] visc: A structure containing vertical viscosities, bottom drag viscosities, and related fields.[in] dirs: A structure containing several relevant directory paths.[inout] ntrunc: A target for the variable that records the number of times the velocity is truncated (this should be 0).
-
subroutine, public mom_dynamics_unsplit_rk2::end_dyn_unsplit_rk2(CS CS) Clean up and deallocate memory associated with the dyn_unsplit_RK2 module.
- Parameters
cs: dyn_unsplit_RK2 control structure that will be deallocated in this subroutine.
-
integer
-
namespace
MOM_energetic_PBL¶ By Robert Hallberg, 2015.
This file contains the subroutine (energetic_PBL) that uses an integrated boundary layer energy budget (like a bulk- or refined- bulk mixed layer scheme), but instead of homogenizing this model calculates a finite diffusivity and viscosity, which in this regard is conceptually similar to what is done with KPP or various two-equation closures. However, the scheme that is implemented here has the big advantage that is entirely implicit, but is simple enough that it requires only a single vertical pass to determine the diffusivity. The development of bulk mixed layer models stems from the work of various people, as described in the review paper by Niiler and Kraus (1979). The work here draws in with particular on the form for TKE decay proposed by Oberhuber (JPO, 1993, 808-829), with an extension to a refined bulk mixed layer as described in Hallberg (Aha Huliko’a, 2003). The physical processes portrayed in this subroutine include convectively driven mixing and mechanically driven mixing. Unlike boundary-layer mixing, stratified shear mixing is not a one-directional turbulent process, and it is dealt with elsewhere in the MOM6 code within the module MOM_kappa_shear.F90. It is assumed that the heat, mass, and salt fluxes have been applied elsewhere, but that their implications for the integrated TKE budget have been captured in an array that is provided as an argument to this subroutine. This is a full 3-d array due to the effects of penetrating shortwave radiation.
-
namespace
mom_energetic_pbl¶ Energetically consistent planetary boundary layer parameterization.
Unnamed Group
-
integer, parameter mom_energetic_pbl::use_fixed_mstar = 0 Enumeration values for mstar_Scheme.
The value of mstar_scheme to use a constant mstar
-
integer, parameter mom_energetic_pbl::mstar_from_ekman = 2 The value of mstar_scheme to base mstar on the ratio of the Ekman layer depth to the Obukhov depth.
-
integer, parameter mom_energetic_pbl::mstar_from_rh18 = 3 The value of mstar_scheme to base mstar of of RH18.
-
integer, parameter mom_energetic_pbl::no_langmuir = 0 The value of LT_ENHANCE_FORM not use Langmuir turbolence.
-
integer, parameter mom_energetic_pbl::langmuir_rescale = 2 The value of LT_ENHANCE_FORM to use a multiplicative rescaling of mstar to account for Langmuir turbulence.
-
integer, parameter mom_energetic_pbl::langmuir_add = 3 The value of LT_ENHANCE_FORM to add a contribution to mstar from Langmuir turblence to other contributions.
-
integer, parameter mom_energetic_pbl::wt_from_croot_tke = 0 Use a constant times the cube root of remaining TKE to calculate the turbulent velocity.
-
integer, parameter mom_energetic_pbl::wt_from_rh18 = 1 Use a scheme based on a combination of w* and v* as documented in Reichl & Hallberg (2018) to calculate the turbulent velocity.
-
character*(20), parameter mom_energetic_pbl::constant_string = "CONSTANT" Enumeration values for mstar_Scheme.
The value of mstar_scheme to use a constant mstar
-
character*(20), parameter mom_energetic_pbl::om4_string = "OM4" Enumeration values for mstar_Scheme.
The value of mstar_scheme to use a constant mstar
-
character*(20), parameter mom_energetic_pbl::rh18_string = "REICHL_H18" Enumeration values for mstar_Scheme.
The value of mstar_scheme to use a constant mstar
-
character*(20), parameter mom_energetic_pbl::root_tke_string = "CUBE_ROOT_TKE" Enumeration values for mstar_Scheme.
The value of mstar_scheme to use a constant mstar
-
character*(20), parameter mom_energetic_pbl::none_string = "NONE" Enumeration values for mstar_Scheme.
The value of mstar_scheme to use a constant mstar
-
character*(20), parameter mom_energetic_pbl::rescaled_string = "RESCALE" Enumeration values for mstar_Scheme.
The value of mstar_scheme to use a constant mstar
-
character*(20), parameter mom_energetic_pbl::additive_string = "ADDITIVE" Enumeration values for mstar_Scheme.
The value of mstar_scheme to use a constant mstar
-
subroutine, public mom_energetic_pbl::energetic_pbl(h_3d h_3d, u_3d u_3d, v_3d v_3d, tv tv, fluxes fluxes, dt dt, Kd_int Kd_int, G G, GV GV, US US, CS CS, dSV_dT dSV_dT, dSV_dS dSV_dS, TKE_forced TKE_forced, buoy_flux buoy_flux, dt_diag dt_diag, last_call last_call, dT_expected dT_expected, dS_expected dS_expected, Waves Waves) This subroutine determines the diffusivities from the integrated energetics mixed layer model. It assumes that heating, cooling and freshwater fluxes have already been applied. All calculations are done implicitly, and there is no stability limit on the time step.
- Parameters
[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[inout] h_3d: Layer thicknesses [H ~> m or kg m-2].[in] u_3d: Zonal velocities interpolated to h points[in] v_3d: Zonal velocities interpolated to h points[in] dsv_dt: The partial derivative of in-situ specific[in] dsv_ds: The partial derivative of in-situ specific[in] tke_forced: The forcing requirements to homogenize the[inout] tv: A structure containing pointers to any available thermodynamic fields. Absent fields have NULL ptrs.[inout] fluxes: A structure containing pointers to any possible forcing fields. Unused fields have NULL ptrs.[in] dt: Time increment [T ~> s].[out] kd_int: The diagnosed diffusivities at interfacescs: The control structure returned by a previous call to mixedlayer_init.[in] buoy_flux: The surface buoyancy flux [Z2 T-3 ~> m2 s-3].[in] dt_diag: The diagnostic time step, which may be less than dt if there are two calls to mixedlayer [T ~> s].[in] last_call: If true, this is the last call to mixedlayer in the current time step, so diagnostics will be written. The default is .true.[out] dt_expected: The values of temperature change that[out] ds_expected: The values of salinity change thatwaves: Wave CS
-
subroutine
epbl_column(h h, u u, v v, T0 T0, S0 S0, dSV_dT dSV_dT, dSV_dS dSV_dS, TKE_forcing TKE_forcing, B_flux B_flux, absf absf, u_star u_star, u_star_mean u_star_mean, dt dt, MLD_io MLD_io, Kd Kd, mixvel mixvel, mixlen mixlen, GV GV, US US, CS CS, eCD eCD, dt_diag dt_diag, Waves Waves, G G, i i, j j)¶ This subroutine determines the diffusivities from the integrated energetics mixed layer model for a single column of water.
- Parameters
[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] h: Layer thicknesses [H ~> m or kg m-2].[in] u: Zonal velocities interpolated to h points [m s-1].[in] v: Zonal velocities interpolated to h points [m s-1].[in] t0: The initial layer temperatures [degC].[in] s0: The initial layer salinities [ppt].[in] dsv_dt: The partial derivative of in-situ specific volume with potential temperature [m3 kg-1 degC-1].[in] dsv_ds: The partial derivative of in-situ specific volume with salinity [m3 kg-1 ppt-1].[in] tke_forcing: The forcing requirements to homogenize the forcing that has been applied to each layer [kg m-3 Z3 T-2 ~> J m-2].[in] b_flux: The surface buoyancy flux [Z2 T-3 ~> m2 s-3][in] absf: The absolute value of the Coriolis parameter [T-1].[in] u_star: The surface friction velocity [Z T-1 ~> m s-1].[in] u_star_mean: The surface friction velocity without any contribution from unresolved gustiness [Z T-1 ~> m s-1].[inout] mld_io: A first guess at the mixed layer depth on input, and the calculated mixed layer depth on output [Z ~> m].[in] dt: Time increment [T ~> s].[out] kd: The diagnosed diffusivities at interfaces[out] mixvel: The mixing velocity scale used in Kd[out] mixlen: The mixing length scale used in Kd [Z ~> m].cs: The control structure returned by a previous call to mixedlayer_init.[inout] ecd: A container for passing around diagnostics.[in] dt_diag: The diagnostic time step, which may be less than dt if there are two calls to mixedlayer [T ~> s].waves: Wave CS for Langmuir turbulence[inout] g: The ocean’s grid structure.[in] i: The i-index to work on (used for Waves)[in] j: The i-index to work on (used for Waves)
-
subroutine
find_pe_chg(Kddt_h0 Kddt_h0, dKddt_h dKddt_h, hp_a hp_a, hp_b hp_b, Th_a Th_a, Sh_a Sh_a, Th_b Th_b, Sh_b Sh_b, dT_to_dPE_a dT_to_dPE_a, dS_to_dPE_a dS_to_dPE_a, dT_to_dPE_b dT_to_dPE_b, dS_to_dPE_b dS_to_dPE_b, pres_Z pres_Z, dT_to_dColHt_a dT_to_dColHt_a, dS_to_dColHt_a dS_to_dColHt_a, dT_to_dColHt_b dT_to_dColHt_b, dS_to_dColHt_b dS_to_dColHt_b, PE_chg PE_chg, dPEc_dKd dPEc_dKd, dPE_max dPE_max, dPEc_dKd_0 dPEc_dKd_0, ColHt_cor ColHt_cor)¶ This subroutine calculates the change in potential energy and or derivatives for several changes in an interfaces’s diapycnal diffusivity times a timestep.
- Parameters
[in] kddt_h0: The previously used diffusivity at an interface times the time step and divided by the average of the thicknesses around the interface [H ~> m or kg m-2].[in] dkddt_h: The trial change in the diffusivity at an interface times the time step and divided by the average of the thicknesses around the interface [H ~> m or kg m-2].[in] hp_a: The effective pivot thickness of the layer above the interface, given by h_k plus a term that is a fraction (determined from the tridiagonal solver) of Kddt_h for the interface above [H ~> m or kg m-2].[in] hp_b: The effective pivot thickness of the layer below the interface, given by h_k plus a term that is a fraction (determined from the tridiagonal solver) of Kddt_h for the interface above [H ~> m or kg m-2].[in] th_a: An effective temperature times a thickness in the layer above, including implicit mixing effects with other yet higher layers [degC H ~> degC m or degC kg m-2].[in] sh_a: An effective salinity times a thickness in the layer above, including implicit mixing effects with other yet higher layers [degC H ~> degC m or degC kg m-2].[in] th_b: An effective temperature times a thickness in the layer below, including implicit mixfing effects with other yet lower layers [degC H ~> degC m or degC kg m-2].[in] sh_b: An effective salinity times a thickness in the layer below, including implicit mixing effects with other yet lower layers [degC H ~> degC m or degC kg m-2].[in] dt_to_dpe_a: A factor (pres_lay*mass_lay*dSpec_vol/dT) relating a layer’s temperature change to the change in column potential energy, including all implicit diffusive changes in the temperatures of all the layers above [kg m-3 Z3 T-2 degC-1 ~> J m-2 degC-1].[in] ds_to_dpe_a: A factor (pres_lay*mass_lay*dSpec_vol/dS) relating a layer’s salinity change to the change in column potential energy, including all implicit diffusive changes in the salinities of all the layers above [kg m-3 Z3 T-2 ppt-1 ~> J m-2 ppt-1].[in] dt_to_dpe_b: A factor (pres_lay*mass_lay*dSpec_vol/dT) relating a layer’s temperature change to the change in column potential energy, including all implicit diffusive changes in the temperatures of all the layers below [kg m-3 Z3 T-2 degC-1 ~> J m-2 degC-1].[in] ds_to_dpe_b: A factor (pres_lay*mass_lay*dSpec_vol/dS) relating a layer’s salinity change to the change in column potential energy, including all implicit diffusive changes in the salinities of all the layers below [kg m-3 Z3 T-2 ppt-1 ~> J m-2 ppt-1].[in] pres_z: The rescaled hydrostatic interface pressure, which relates the changes in column thickness to the energy that is radiated as gravity waves and unavailable to drive mixing [J m-2 Z-1 ~> J m-3].[in] dt_to_dcolht_a: A factor (mass_lay*dSColHtc_vol/dT) relating a layer’s temperature change to the change in column height, including all implicit diffusive changes in the temperatures of all the layers above [Z degC-1 ~> m degC-1].[in] ds_to_dcolht_a: A factor (mass_lay*dSColHtc_vol/dS) relating a layer’s salinity change to the change in column height, including all implicit diffusive changes in the salinities of all the layers above [Z ppt-1 ~> m ppt-1].[in] dt_to_dcolht_b: A factor (mass_lay*dSColHtc_vol/dT) relating a layer’s temperature change to the change in column height, including all implicit diffusive changes in the temperatures of all the layers below [Z degC-1 ~> m degC-1].[in] ds_to_dcolht_b: A factor (mass_lay*dSColHtc_vol/dS) relating a layer’s salinity change to the change in column height, including all implicit diffusive changes in the salinities of all the layers below [Z ppt-1 ~> m ppt-1].[out] pe_chg: The change in column potential energy from applying Kddt_h at the present interface [kg m-3 Z3 T-2 ~> J m-2].[out] dpec_dkd: The partial derivative of PE_chg with Kddt_h [J m-2 H-1 ~> J m-3 or J kg-1].[out] dpe_max: The maximum change in column potential energy that could be realizedd by applying a huge value of Kddt_h at the present interface [kg m-3 Z3 T-2 ~> J m-2].[out] dpec_dkd_0: The partial derivative of PE_chg with Kddt_h in the limit where Kddt_h = 0 [kg m-3 Z3 T-2 H-1 ~> J m-3 or J kg-1].[out] colht_cor: The correction to PE_chg that is made due to a net change in the column height [kg m-3 Z3 T-2 ~> J m-2].
-
subroutine
find_pe_chg_orig(Kddt_h Kddt_h, h_k h_k, b_den_1 b_den_1, dTe_term dTe_term, dSe_term dSe_term, dT_km1_t2 dT_km1_t2, dS_km1_t2 dS_km1_t2, dT_to_dPE_k dT_to_dPE_k, dS_to_dPE_k dS_to_dPE_k, dT_to_dPEa dT_to_dPEa, dS_to_dPEa dS_to_dPEa, pres_Z pres_Z, dT_to_dColHt_k dT_to_dColHt_k, dS_to_dColHt_k dS_to_dColHt_k, dT_to_dColHta dT_to_dColHta, dS_to_dColHta dS_to_dColHta, PE_chg PE_chg, dPEc_dKd dPEc_dKd, dPE_max dPE_max, dPEc_dKd_0 dPEc_dKd_0)¶ This subroutine calculates the change in potential energy and or derivatives for several changes in an interfaces’s diapycnal diffusivity times a timestep using the original form used in the first version of ePBL.
- Parameters
[in] kddt_h: The diffusivity at an interface times the time step and divided by the average of the thicknesses around the interface [H ~> m or kg m-2].[in] h_k: The thickness of the layer below the interface [H ~> m or kg m-2].[in] b_den_1: The first term in the denominator of the pivot for the tridiagonal solver, given by h_k plus a term that is a fraction (determined from the tridiagonal solver) of Kddt_h for the interface above [H ~> m or kg m-2].[in] dte_term: A diffusivity-independent term related to the temperature change in the layer below the interface [degC H ~> degC m or degC kg m-2].[in] dse_term: A diffusivity-independent term related to the salinity change in the layer below the interface [ppt H ~> ppt m or ppt kg m-2].[in] dt_km1_t2: A diffusivity-independent term related to the temperature change in the layer above the interface [degC].[in] ds_km1_t2: A diffusivity-independent term related to the salinity change in the layer above the interface [ppt].[in] pres_z: The rescaled hydrostatic interface pressure, which relates the changes in column thickness to the energy that is radiated as gravity waves and unavailable to drive mixing [J m-2 Z-1 ~> J m-3].[in] dt_to_dpe_k: A factor (pres_lay*mass_lay*dSpec_vol/dT) relating a layer’s temperature change to the change in column potential energy, including all implicit diffusive changes in the temperatures of all the layers below [kg m-3 Z3 T-2 degC-1 ~> J m-2 degC-1].[in] ds_to_dpe_k: A factor (pres_lay*mass_lay*dSpec_vol/dS) relating a layer’s salinity change to the change in column potential energy, including all implicit diffusive changes in the in the salinities of all the layers below [kg m-3 Z3 T-2 ppt-1 ~> J m-2 ppt-1].[in] dt_to_dpea: A factor (pres_lay*mass_lay*dSpec_vol/dT) relating a layer’s temperature change to the change in column potential energy, including all implicit diffusive changes in the temperatures of all the layers above [kg m-3 Z3 T-2 degC-1 ~> J m-2 degC-1].[in] ds_to_dpea: A factor (pres_lay*mass_lay*dSpec_vol/dS) relating a layer’s salinity change to the change in column potential energy, including all implicit diffusive changes in the salinities of all the layers above [kg m-3 Z3 T-2 ppt-1 ~> J m-2 ppt-1].[in] dt_to_dcolht_k: A factor (mass_lay*dSColHtc_vol/dT) relating a layer’s temperature change to the change in column height, including all implicit diffusive changes in the temperatures of all the layers below [Z degC-1 ~> m degC-1].[in] ds_to_dcolht_k: A factor (mass_lay*dSColHtc_vol/dS) relating a layer’s salinity change to the change in column height, including all implicit diffusive changes in the salinities of all the layers below [Z ppt-1 ~> m ppt-1].[in] dt_to_dcolhta: A factor (mass_lay*dSColHtc_vol/dT) relating a layer’s temperature change to the change in column height, including all implicit diffusive changes in the temperatures of all the layers above [Z degC-1 ~> m degC-1].[in] ds_to_dcolhta: A factor (mass_lay*dSColHtc_vol/dS) relating a layer’s salinity change to the change in column height, including all implicit diffusive changes in the salinities of all the layers above [Z ppt-1 ~> m ppt-1].[out] pe_chg: The change in column potential energy from applying Kddt_h at the present interface [kg m-3 Z3 T-2 ~> J m-2].[out] dpec_dkd: The partial derivative of PE_chg with Kddt_h [kg m-3 Z3 T-2 H-1 ~> J m-3 or J kg-1].[out] dpe_max: The maximum change in column potential energy that could be realizedd by applying a huge value of Kddt_h at the present interface [kg m-3 Z3 T-2 ~> J m-2].[out] dpec_dkd_0: The partial derivative of PE_chg with Kddt_h in the limit where Kddt_h = 0 [kg m-3 Z3 T-2 H-1 ~> J m-3 or J kg-1].
-
subroutine
find_mstar(CS CS, US US, Buoyancy_Flux Buoyancy_Flux, UStar UStar, UStar_Mean UStar_Mean, BLD BLD, Abs_Coriolis Abs_Coriolis, MStar MStar, Langmuir_Number Langmuir_Number, MStar_LT MStar_LT, Convect_Langmuir_Number Convect_Langmuir_Number)¶ This subroutine finds the Mstar value for ePBL.
- Parameters
cs: Energetic_PBL control structure.[in] us: A dimensional unit scaling type[in] ustar: ustar w/ gustiness [Z T-1 ~> m s-1][in] ustar_mean: ustar w/o gustiness [Z T-1 ~> m s-1][in] abs_coriolis: abolute value of the Coriolis parameter [T-1][in] buoyancy_flux: Buoyancy flux [Z2 T-3 ~> m2 s-3][in] bld: boundary layer depth [Z ~> m][out] mstar: Ouput mstar (Mixing/ustar**3) [nondim][in] langmuir_number: Langmuir number [nondim][out] mstar_lt: Mstar increase due to Langmuir turbulence [nondim][out] convect_langmuir_number: Langmuir number including buoyancy flux [nondim]
-
subroutine
mstar_langmuir(CS CS, US US, abs_Coriolis abs_Coriolis, buoyancy_flux buoyancy_flux, ustar ustar, BLD BLD, Langmuir_Number Langmuir_Number, mstar mstar, mstar_LT mstar_LT, Convect_Langmuir_Number Convect_Langmuir_Number)¶ This subroutine modifies the Mstar value if the Langmuir number is present.
- Parameters
cs: Energetic_PBL control structure.[in] us: A dimensional unit scaling type[in] abs_coriolis: Absolute value of the Coriolis parameter [T-1 ~> s-1][in] buoyancy_flux: Buoyancy flux [Z2 T-3 ~> m2 s-3][in] ustar: Surface friction velocity with? gustiness [Z T-1 ~> m s-1][in] bld: boundary layer depth [Z ~> m][inout] mstar: Input/output mstar (Mixing/ustar**3) [nondim][in] langmuir_number: Langmuir number [nondim][out] mstar_lt: Mstar increase due to Langmuir turbulence [nondim][out] convect_langmuir_number: Langmuir number including buoyancy flux [nondim]
-
subroutine, public mom_energetic_pbl::energetic_pbl_get_mld(CS CS, MLD MLD, G G, US US, m_to_MLD_units m_to_MLD_units) Copies the ePBL active mixed layer depth into MLD.
- Parameters
cs: Control structure for ePBL[in] g: Grid structure[in] us: A dimensional unit scaling type[out] mld: Depth of ePBL active mixing layer [m or other units][in] m_to_mld_units: A conversion factor to the desired units for MLD
-
subroutine, public mom_energetic_pbl::energetic_pbl_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS) This subroutine initializes the energetic_PBL module.
- Parameters
[in] time: The current model time[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters[inout] diag: A structure that is used to regulate diagnostic outputcs: A pointer that is set to point to the control structure for this module
-
subroutine, public mom_energetic_pbl::energetic_pbl_end(CS CS) Clean up and deallocate memory associated with the energetic_PBL module.
- Parameters
cs: Energetic_PBL control structure that will be deallocated in this subroutine.
-
-
namespace
mom_entrain_diffusive¶ Diapycnal mixing and advection in isopycnal mode.
By Robert Hallberg, September 1997 - July 2000
This file contains the subroutines that implement diapycnal mixing and advection in isopycnal layers. The main subroutine, calculate_entrainment, returns the entrainment by each layer across the interfaces above and below it. These are calculated subject to the constraints that no layers can be driven to neg- ative thickness and that the each layer maintains its target density, using the scheme described in Hallberg (MWR 2000). There may or may not be a bulk mixed layer above the isopycnal layers. The solution is iterated until the change in the entrainment between successive iterations is less than some small tolerance.
The dual-stream entrainment scheme of MacDougall and Dewar (JPO 1997) is used for combined diapycnal advection and diffusion, modified as described in Hallberg (MWR 2000) to be solved implicitly in time. Any profile of diffusivities may be used. Diapycnal advection is fundamentally the residual of diapycnal diffusion, so the fully implicit upwind differencing scheme that is used is entirely appropriate. The downward buoyancy flux in each layer is determined from an implicit calculation based on the previously calculated flux of the layer above and an estim- ated flux in the layer below. This flux is subject to the foll- owing conditions: (1) the flux in the top and bottom layers are set by the boundary conditions, and (2) no layer may be driven below an Angstrom thickness. If there is a bulk mixed layer, the mixed and buffer layers are treated as Eulerian layers, whose thicknesses only change due to entrainment by the interior layers.
Functions
-
subroutine, public mom_entrain_diffusive::entrainment_diffusive(h h, tv tv, fluxes fluxes, dt dt, G G, GV GV, US US, CS CS, ea ea, eb eb, kb_out kb_out, Kd_Lay Kd_Lay, Kd_int Kd_int) This subroutine calculates ea and eb, the rates at which a layer entrains from the layers above and below. The entrainment rates are proportional to the buoyancy flux in a layer and inversely proportional to the density differences between layers. The scheme that is used here is described in detail in Hallberg, Mon. Wea. Rev. 2000.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] h: Layer thicknesses [H ~> m or kg m-2].[in] tv: A structure containing pointers to any available thermodynamic fields. Absent fields have NULL ptrs.[in] fluxes: A structure of surface fluxes that may be used.[in] dt: The time increment [T ~> s].cs: The control structure returned by a previous call to entrain_diffusive_init.[out] ea: The amount of fluid entrained from the layer[out] eb: The amount of fluid entrained from the layer[inout] kb_out: The index of the lightest layer denser than[in] kd_lay: The diapycnal diffusivity of layers[in] kd_int: The diapycnal diffusivity of interfaces
-
subroutine
f_to_ent(F F, h h, kb kb, kmb kmb, j j, G G, GV GV, CS CS, dsp1_ds dsp1_ds, eakb eakb, Ent_bl Ent_bl, ea ea, eb eb, do_i_in do_i_in)¶ This subroutine calculates the actual entrainments (ea and eb) and the amount of surface forcing that is applied to each layer if there is no bulk mixed layer.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] f: The density flux through a layer within a time step divided by the density difference across the interface below the layer [H ~> m or kg m-2].[in] h: Layer thicknesses [H ~> m or kg m-2][in] kb: The index of the lightest layer denser than the deepest buffer layer.[in] kmb: The number of mixed and buffer layers.[in] j: The meridional index upon which to work.[in] cs: This module’s control structure.[in] dsp1_ds: The ratio of coordinate variable differences across the interfaces below a layer over the difference across the interface above the layer.[in] eakb: The entrainment from above by the layer below the buffer layer [H ~> m or kg m-2].[in] ent_bl: The average entrainment upward and downward across each interface around the buffer layers [H ~> m or kg m-2].[inout] ea: The amount of fluid entrained from the layer[inout] eb: The amount of fluid entrained from the layer[in] do_i_in: Indicates which i-points to work on.
-
subroutine
set_ent_bl(h h, dtKd_int dtKd_int, tv tv, kb kb, kmb kmb, do_i do_i, G G, GV GV, CS CS, j j, Ent_bl Ent_bl, Sref Sref, h_bl h_bl)¶ This subroutine sets the average entrainment across each of the interfaces between buffer layers within a timestep. It also causes thin and relatively light interior layers to be entrained by the deepest buffer layer. Also find the initial coordinate potential densities (Sref) of each layer.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] h: Layer thicknesses [H ~> m or kg m-2][in] dtkd_int: The diapycnal diffusivity across[in] tv: A structure containing pointers to any available thermodynamic fields. Absent fields have NULL ptrs.[inout] kb: The index of the lightest layer denser than the buffer layer or 1 if there is no buffer layer.[in] kmb: The number of mixed and buffer layers.[in] do_i: A logical variable indicating which i-points to work on.cs: This module’s control structure.[in] j: The meridional index upon which to work.[out] ent_bl: The average entrainment upward and[out] sref: The coordinate potential density minus 1000 for each layer [kg m-3].[out] h_bl: The thickness of each layer [H ~> m or kg m-2].
-
subroutine
determine_dskb(h_bl h_bl, Sref Sref, Ent_bl Ent_bl, E_kb E_kb, is is, ie ie, kmb kmb, G G, GV GV, limit limit, dSkb dSkb, ddSkb_dE ddSkb_dE, dSlay dSlay, ddSlay_dE ddSlay_dE, dS_anom_lim dS_anom_lim, do_i_in do_i_in)¶ This subroutine determines the reference density difference between the bottommost buffer layer and the first interior after the mixing between mixed and buffer layers and mixing with the layer below. Within the mixed and buffer layers, entrainment from the layer above is increased when it is necessary to keep the layers from developing a negative thickness; otherwise it equals Ent_bl. At each interface, the upward and downward fluxes average out to Ent_bl, unless entrainment by the layer below is larger than twice Ent_bl. The density difference across the first interior layer may also be returned. It could also be limited to avoid negative values or values that greatly exceed the density differences across an interface. Additionally, the partial derivatives of dSkb and dSlay with E_kb could also be returned.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] h_bl: Layer thickness [H ~> m or kg m-2][in] sref: Reference potential density [kg m-3][in] ent_bl: The average entrainment upward and downward across each interface around the buffer layers [H ~> m or kg m-2].[in] e_kb: The entrainment by the top interior layer [H ~> m or kg m-2].[in] is: The start of the i-index range to work on.[in] ie: The end of the i-index range to work on.[in] kmb: The number of mixed and buffer layers.[in] limit: If true, limit dSkb and dSlay to avoid negative values.[inout] dskb: The limited potential density difference across the interface between the bottommost buffer layer and the topmost interior layer. dSkb > 0.[inout] ddskb_de: The partial derivative of dSkb with E [kg m-3 H-1 ~> kg m-4 or m-1].[inout] dslay: The limited potential density difference across the topmost interior layer. 0 < dSkb[inout] ddslay_de: The partial derivative of dSlay with E [kg m-3 H-1 ~> kg m-4 or m-1].[inout] ds_anom_lim: A limiting value to use for the density anomalies below the buffer layer [kg m-3].[in] do_i_in: If present, determines which columns are worked on.
-
subroutine
f_kb_to_ea_kb(h_bl h_bl, Sref Sref, Ent_bl Ent_bl, I_dSkbp1 I_dSkbp1, F_kb F_kb, kmb kmb, i i, G G, GV GV, CS CS, ea_kb ea_kb, tol_in tol_in)¶ Given an entrainment from below for layer kb, determine a consistent entrainment from above, such that dSkb * ea_kb = dSkbp1 * F_kb. The input value of ea_kb is both the maximum value that can be obtained and the first guess of the iterations. Ideally ea_kb should be an under-estimate.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h_bl: Layer thickness, with the top interior[in] sref: The coordinate reference potential density,[in] ent_bl: The average entrainment upward and downward[in] i_dskbp1: The inverse of the difference in reference potential density across the base of the uppermost interior layer [m3 kg-1].[in] f_kb: The entrainment from below by the uppermost interior layer [H ~> m or kg m-2][in] kmb: The number of mixed and buffer layers.[in] i: The i-index to work oncs: This module’s control structure.[inout] ea_kb: The entrainment from above by the layer below the buffer layer (i.e. layer kb) [H ~> m or kg m-2].[in] tol_in: A tolerance for the iterative determination of the entrainment [H ~> m or kg m-2].
-
subroutine
determine_ea_kb(h_bl h_bl, dtKd_kb dtKd_kb, Sref Sref, I_dSkbp1 I_dSkbp1, Ent_bl Ent_bl, ea_kbp1 ea_kbp1, min_eakb min_eakb, max_eakb max_eakb, kmb kmb, is is, ie ie, do_i do_i, G G, GV GV, CS CS, Ent Ent, error error, err_min_eakb0 err_min_eakb0, err_max_eakb0 err_max_eakb0, F_kb F_kb, dFdfm_kb dFdfm_kb)¶ This subroutine determines the entrainment from above by the top interior layer (labeled kb elsewhere) given an entrainment by the layer below it, constrained to be within the provided bounds.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] h_bl: Layer thickness, with the top interior layer at k-index kmb+1 [H ~> m or kg m-2].[in] sref: The coordinate reference potential density, with the value of the topmost interior layer at layer kmb+1 [kg m-3].[in] ent_bl: The average entrainment upward and downward across each interface around the buffer layers [H ~> m or kg m-2].[in] i_dskbp1: The inverse of the difference in reference potential density across the base of the uppermost interior layer [m3 kg-1].[in] dtkd_kb: The diapycnal diffusivity in the top interior layer times the time step [H2 ~> m2 or kg2 m-4].[in] ea_kbp1: The entrainment from above by layer kb+1 [H ~> m or kg m-2].[in] min_eakb: The minimum permissible rate of entrainment [H ~> m or kg m-2].[in] max_eakb: The maximum permissible rate of entrainment [H ~> m or kg m-2].[in] kmb: The number of mixed and buffer layers.[in] is: The start of the i-index range to work on.[in] ie: The end of the i-index range to work on.[in] do_i: A logical variable indicating which i-points to work on.cs: This module’s control structure.[inout] ent: The entrainment rate of the uppermost interior layer [H ~> m or kg m-2]. The input value is the first guess.[out] error: The error (locally defined in this routine) associated with the returned solution.[in] err_min_eakb0: The errors (locally defined) associated with min_eakb when ea_kbp1 = 0, returned from a previous call to this fn.[in] err_max_eakb0: The errors (locally defined) associated with min_eakb when ea_kbp1 = 0, returned from a previous call to this fn.[out] f_kb: The entrainment from below by the uppermost interior layer corresponding to the returned value of Ent [H ~> m or kg m-2].[out] dfdfm_kb: The partial derivative of F_kb with ea_kbp1 [nondim].
-
subroutine
find_maxf_kb(h_bl h_bl, Sref Sref, Ent_bl Ent_bl, I_dSkbp1 I_dSkbp1, min_ent_in min_ent_in, max_ent_in max_ent_in, kmb kmb, is is, ie ie, G G, GV GV, CS CS, maxF maxF, ent_maxF ent_maxF, do_i_in do_i_in, F_lim_maxent F_lim_maxent, F_thresh F_thresh)¶ Maximize F = ent*ds_kb*I_dSkbp1 in the range min_ent < ent < max_ent.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] h_bl: Layer thickness [H ~> m or kg m-2][in] sref: Reference potential density [kg m-3].[in] ent_bl: The average entrainment upward and[in] i_dskbp1: The inverse of the difference in reference potential density across the base of the uppermost interior layer [m3 kg-1].[in] min_ent_in: The minimum value of ent to search, [H ~> m or kg m-2].[in] max_ent_in: The maximum value of ent to search, [H ~> m or kg m-2].[in] kmb: The number of mixed and buffer layers.[in] is: The start of the i-index range to work on.[in] ie: The end of the i-index range to work on.cs: This module’s control structure.[out] maxf: The maximum value of F = ent*ds_kb*I_dSkbp1 found in the range min_ent < ent < max_ent [H ~> m or kg m-2].[out] ent_maxf: The value of ent at that maximum [H ~> m or kg m-2].[in] do_i_in: A logical array indicating which columns[out] f_lim_maxent: If present, do not apply the limit in[in] f_thresh: If F_thresh is present, return the first
-
subroutine, public mom_entrain_diffusive::entrain_diffusive_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS) This subroutine initializes the parameters and memory associated with the entrain_diffusive module.
- Parameters
[in] time: The current model time.[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters.[inout] diag: A structure that is used to regulate diagnostic output.cs: A pointer that is set to point to the control structure.
-
subroutine, public mom_entrain_diffusive::entrain_diffusive_end(CS CS) This subroutine cleans up and deallocates any memory associated with the entrain_diffusive module.
- Parameters
cs: A pointer to the control structure for this module that will be deallocated.
-
-
namespace
mom_eos¶ Provides subroutines for quantities specific to the equation of state.
The MOM_EOS module is a wrapper for various equations of state (e.g. Linear, Wright, UNESCO) and provides a uniform interface to the rest of the model independent of which equation of state is being used.
Functions
-
subroutine
calculate_density_scalar(T T, S S, pressure pressure, rho rho, EOS EOS, rho_ref rho_ref)¶ Calls the appropriate subroutine to calculate density of sea water for scalar inputs. If rho_ref is present, the anomaly with respect to rho_ref is returned.
- Parameters
[in] t: Potential temperature referenced to the surface [degC][in] s: Salinity [ppt][in] pressure: Pressure [Pa][out] rho: Density (in-situ if pressure is local) [kg m-3]eos: Equation of state structure[in] rho_ref: A reference density [kg m-3].
-
subroutine
calculate_density_array(T T, S S, pressure pressure, rho rho, start start, npts npts, EOS EOS, rho_ref rho_ref)¶ Calls the appropriate subroutine to calculate the density of sea water for 1-D array inputs. If rho_ref is present, the anomaly with respect to rho_ref is returned.
- Parameters
[in] t: Potential temperature referenced to the surface [degC][in] s: Salinity [ppt][in] pressure: Pressure [Pa][out] rho: Density (in-situ if pressure is local) [kg m-3][in] start: Start index for computation[in] npts: Number of point to computeeos: Equation of state structure[in] rho_ref: A reference density [kg m-3].
-
subroutine
calculate_spec_vol_scalar(T T, S S, pressure pressure, specvol specvol, EOS EOS, spv_ref spv_ref)¶ Calls the appropriate subroutine to calculate specific volume of sea water for scalar inputs.
- Parameters
[in] t: Potential temperature referenced to the surface [degC][in] s: Salinity [ppt][in] pressure: Pressure [Pa][out] specvol: specific volume (in-situ if pressure is local) [m3 kg-1]eos: Equation of state structure[in] spv_ref: A reference specific volume [m3 kg-1].
-
subroutine
calculate_spec_vol_array(T T, S S, pressure pressure, specvol specvol, start start, npts npts, EOS EOS, spv_ref spv_ref)¶ Calls the appropriate subroutine to calculate the specific volume of sea water for 1-D array inputs.
- Parameters
[in] t: potential temperature relative to the surface [degC].[in] s: salinity [ppt].[in] pressure: pressure [Pa].[out] specvol: in situ specific volume [kg m-3].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.eos: Equation of state structure[in] spv_ref: A reference specific volume [m3 kg-1].
-
subroutine
calculate_tfreeze_scalar(S S, pressure pressure, T_fr T_fr, EOS EOS)¶ Calls the appropriate subroutine to calculate the freezing point for scalar inputs.
- Parameters
[in] s: Salinity [ppt][in] pressure: Pressure [Pa][out] t_fr: Freezing point potential temperature referenced to the surface [degC]eos: Equation of state structure
-
subroutine
calculate_tfreeze_array(S S, pressure pressure, T_fr T_fr, start start, npts npts, EOS EOS)¶ Calls the appropriate subroutine to calculate the freezing point for a 1-D array.
- Parameters
[in] s: Salinity [ppt][in] pressure: Pressure [Pa][out] t_fr: Freezing point potential temperature referenced to the surface [degC][in] start: Starting index within the array[in] npts: The number of values to calculateeos: Equation of state structure
-
subroutine
calculate_density_derivs_array(T T, S S, pressure pressure, drho_dT drho_dT, drho_dS drho_dS, start start, npts npts, EOS EOS)¶ Calls the appropriate subroutine to calculate density derivatives for 1-D array inputs.
- Parameters
[in] t: Potential temperature referenced to the surface [degC][in] s: Salinity [ppt][in] pressure: Pressure [Pa][out] drho_dt: The partial derivative of density with potential temperature [kg m-3 degC-1].[out] drho_ds: The partial derivative of density with salinity, in [kg m-3 ppt-1].[in] start: Starting index within the array[in] npts: The number of values to calculateeos: Equation of state structure
-
subroutine
calculate_density_derivs_scalar(T T, S S, pressure pressure, drho_dT drho_dT, drho_dS drho_dS, EOS EOS)¶ Calls the appropriate subroutines to calculate density derivatives by promoting a scalar to a one-element array.
- Parameters
[in] t: Potential temperature referenced to the surface [degC][in] s: Salinity [ppt][in] pressure: Pressure [Pa][out] drho_dt: The partial derivative of density with potential temperature [kg m-3 degC-1].[out] drho_ds: The partial derivative of density with salinity, in [kg m-3 ppt-1].eos: Equation of state structure
-
subroutine
calculate_density_second_derivs_array(T T, S S, pressure pressure, drho_dS_dS drho_dS_dS, drho_dS_dT drho_dS_dT, drho_dT_dT drho_dT_dT, drho_dS_dP drho_dS_dP, drho_dT_dP drho_dT_dP, start start, npts npts, EOS EOS)¶ Calls the appropriate subroutine to calculate density second derivatives for 1-D array inputs.
- Parameters
[in] t: Potential temperature referenced to the surface [degC][in] s: Salinity [ppt][in] pressure: Pressure [Pa][out] drho_ds_ds: Partial derivative of beta with respect to S [kg m-3 ppt-2][out] drho_ds_dt: Partial derivative of beta with respcct to T [kg m-3 ppt-1 degC-1][out] drho_dt_dt: Partial derivative of alpha with respect to T [kg m-3 degC-2][out] drho_ds_dp: Partial derivative of beta with respect to pressure [kg m-3 ppt-1 Pa-1][out] drho_dt_dp: Partial derivative of alpha with respect to pressure [kg m-3 degC-1 Pa-1][in] start: Starting index within the array[in] npts: The number of values to calculateeos: Equation of state structure
-
subroutine
calculate_density_second_derivs_scalar(T T, S S, pressure pressure, drho_dS_dS drho_dS_dS, drho_dS_dT drho_dS_dT, drho_dT_dT drho_dT_dT, drho_dS_dP drho_dS_dP, drho_dT_dP drho_dT_dP, EOS EOS)¶ Calls the appropriate subroutine to calculate density second derivatives for scalar nputs.
- Parameters
[in] t: Potential temperature referenced to the surface [degC][in] s: Salinity [ppt][in] pressure: Pressure [Pa][out] drho_ds_ds: Partial derivative of beta with respect to S [kg m-3 ppt-2][out] drho_ds_dt: Partial derivative of beta with respcct to T [kg m-3 ppt-1 degC-1][out] drho_dt_dt: Partial derivative of alpha with respect to T [kg m-3 degC-2][out] drho_ds_dp: Partial derivative of beta with respect to pressure [kg m-3 ppt-1 Pa-1][out] drho_dt_dp: Partial derivative of alpha with respect to pressure [kg m-3 degC-1 Pa-1]eos: Equation of state structure
-
subroutine, public mom_eos::calculate_specific_vol_derivs(T T, S S, pressure pressure, dSV_dT dSV_dT, dSV_dS dSV_dS, start start, npts npts, EOS EOS) Calls the appropriate subroutine to calculate specific volume derivatives for an array.
- Parameters
[in] t: Potential temperature referenced to the surface [degC][in] s: Salinity [ppt][in] pressure: Pressure [Pa][out] dsv_dt: The partial derivative of specific volume with potential temperature [m3 kg-1 degC-1].[out] dsv_ds: The partial derivative of specific volume with salinity [m3 kg-1 ppt-1].[in] start: Starting index within the array[in] npts: The number of values to calculateeos: Equation of state structure
-
subroutine, public mom_eos::calculate_compress(T T, S S, pressure pressure, rho rho, drho_dp drho_dp, start start, npts npts, EOS EOS) Calls the appropriate subroutine to calculate the density and compressibility for 1-D array inputs.
- Parameters
[in] t: Potential temperature referenced to the surface [degC][in] s: Salinity [ppt][in] pressure: Pressure [Pa][out] rho: In situ density [kg m-3].[out] drho_dp: The partial derivative of density with pressure (also the inverse of the square of sound speed) in s2 m-2.[in] start: Starting index within the array[in] npts: The number of values to calculateeos: Equation of state structure
-
subroutine, public mom_eos::int_specific_vol_dp(T T, S S, p_t p_t, p_b p_b, alpha_ref alpha_ref, HI HI, EOS EOS, dza dza, intp_dza intp_dza, intx_dza intx_dza, inty_dza inty_dza, halo_size halo_size, bathyP bathyP, dP_tiny dP_tiny, useMassWghtInterp useMassWghtInterp) Calls the appropriate subroutine to alculate analytical and nearly-analytical integrals in pressure across layers of geopotential anomalies, which are required for calculating the finite-volume form pressure accelerations in a non-Boussinesq model. There are essentially no free assumptions, apart from the use of Bode’s rule to do the horizontal integrals, and from a truncation in the series for log(1-eps/1+eps) that assumes that |eps| < .
- Parameters
[in] hi: The horizontal index structure[in] t: Potential temperature referenced to the surface [degC][in] s: Salinity [ppt][in] p_t: Pressure at the top of the layer [Pa].[in] p_b: Pressure at the bottom of the layer [Pa].[in] alpha_ref: A mean specific volume that is subtracted out to reduce the magnitude of each of the integrals, m3 kg-1. The calculation is mathematically identical with different values of alpha_ref, but this reduces the effects of roundoff.eos: Equation of state structure[out] dza: The change in the geopotential anomaly across[out] intp_dza: The integral in pressure through the layer of the[out] intx_dza: The integral in x of the difference between the[out] inty_dza: The integral in y of the difference between the[in] halo_size: The width of halo points on which to calculate dza.[in] bathyp: The pressure at the bathymetry [Pa][in] dp_tiny: A miniscule pressure change with the same units as p_t (Pa?)[in] usemasswghtinterp: If true, uses mass weighting to interpolate T/S for top and bottom integrals.
-
subroutine, public mom_eos::int_density_dz(T T, S S, z_t z_t, z_b z_b, rho_ref rho_ref, rho_0 rho_0, G_e G_e, HII HII, HIO HIO, EOS EOS, dpa dpa, intz_dpa intz_dpa, intx_dpa intx_dpa, inty_dpa inty_dpa, bathyT bathyT, dz_neglect dz_neglect, useMassWghtInterp useMassWghtInterp) This subroutine calculates analytical and nearly-analytical integrals of pressure anomalies across layers, which are required for calculating the finite-volume form pressure accelerations in a Boussinesq model.
- Parameters
[in] hii: Ocean horizontal index structures for the input arrays[in] hio: Ocean horizontal index structures for the output arrays[in] t: Potential temperature referenced to the surface [degC][in] s: Salinity [ppt][in] z_t: Height at the top of the layer in depth units [Z ~> m].[in] z_b: Height at the bottom of the layer [Z ~> m].[in] rho_ref: A mean density [kg m-3], that is subtracted out to reduce the magnitude of each of the integrals.[in] rho_0: A density [kg m-3], that is used to calculate the pressure (as p~=-z*rho_0*G_e) used in the equation of state.[in] g_e: The Earth’s gravitational acceleration [m2 Z-1 s-2 ~> m s-2].eos: Equation of state structure[out] dpa: The change in the pressure anomaly across the layer [Pa].[out] intz_dpa: The integral through the thickness of the layer of[out] intx_dpa: The integral in x of the difference between the[out] inty_dpa: The integral in y of the difference between the[in] bathyt: The depth of the bathymetry [Z ~> m].[in] dz_neglect: A miniscule thickness change [Z ~> m].[in] usemasswghtinterp: If true, uses mass weighting to interpolate T/S for top and bottom integrals.
-
logical function, public mom_eos::query_compressible(EOS EOS) Returns true if the equation of state is compressible (i.e. has pressure dependence)
- Parameters
eos: Equation of state structure
-
subroutine, public mom_eos::eos_init(param_file param_file, EOS EOS) Initializes EOS_type by allocating and reading parameters.
- Parameters
[in] param_file: Parameter file structureeos: Equation of state structure
-
subroutine, public mom_eos::eos_manual_init(EOS EOS, form_of_EOS form_of_EOS, form_of_TFreeze form_of_TFreeze, EOS_quadrature EOS_quadrature, Compressible Compressible, Rho_T0_S0 Rho_T0_S0, drho_dT drho_dT, dRho_dS dRho_dS, TFr_S0_P0 TFr_S0_P0, dTFr_dS dTFr_dS, dTFr_dp dTFr_dp) Manually initialized an EOS type (intended for unit testing of routines which need a specific EOS)
- Parameters
eos: Equation of state structure[in] form_of_eos: A coded integer indicating the equation of state to use.[in] form_of_tfreeze: A coded integer indicating the expression for the potential temperature of the freezing point.[in] eos_quadrature: If true, always use the generic (quadrature) code for the integrals of density.[in] compressible: If true, in situ density is a function of pressure.[in] rho_t0_s0: Density at T=0 degC and S=0 ppt [kg m-3][in] drho_dt: Partial derivative of density with temperature in [kg m-3 degC-1][in] drho_ds: Partial derivative of density with salinity in [kg m-3 ppt-1][in] tfr_s0_p0: The freezing potential temperature at S=0, P=0 [degC].[in] dtfr_ds: The derivative of freezing point with salinity in [degC ppt-1].[in] dtfr_dp: The derivative of freezing point with pressure in [degC Pa-1].
-
subroutine, public mom_eos::eos_allocate(EOS EOS) Allocates EOS_type.
- Parameters
eos: Equation of state structure
-
subroutine, public mom_eos::eos_end(EOS EOS) Deallocates EOS_type.
- Parameters
eos: Equation of state structure
-
subroutine, public mom_eos::eos_use_linear(Rho_T0_S0 Rho_T0_S0, dRho_dT dRho_dT, dRho_dS dRho_dS, EOS EOS, use_quadrature use_quadrature) Set equation of state structure (EOS) to linear with given coefficients.
- Note
This routine is primarily for testing and allows a local copy of the EOS_type (EOS argument) to be set to use the linear equation of state independent from the rest of the model.
- Parameters
[in] rho_t0_s0: Density at T=0 degC and S=0 ppt [kg m-3][in] drho_dt: Partial derivative of density with temperature [kg m-3 degC-1][in] drho_ds: Partial derivative of density with salinity [kg m-3 ppt-1][in] use_quadrature: If true, always use the generic (quadrature) code for the integrals of density.eos: Equation of state structure
-
subroutine, public mom_eos::int_density_dz_generic(T T, S S, z_t z_t, z_b z_b, rho_ref rho_ref, rho_0 rho_0, G_e G_e, HII HII, HIO HIO, EOS EOS, dpa dpa, intz_dpa intz_dpa, intx_dpa intx_dpa, inty_dpa inty_dpa, bathyT bathyT, dz_neglect dz_neglect, useMassWghtInterp useMassWghtInterp) This subroutine calculates (by numerical quadrature) integrals of pressure anomalies across layers, which are required for calculating the finite-volume form pressure accelerations in a Boussinesq model.
- Parameters
[in] hii: Horizontal index type for input variables.[in] hio: Horizontal index type for output variables.[in] t: Potential temperature of the layer [degC].[in] s: Salinity of the layer [ppt].[in] z_t: Height at the top of the layer in depth units [Z ~> m].[in] z_b: Height at the bottom of the layer [Z ~> m].[in] rho_ref: A mean density [kg m-3], that is subtracted out to reduce the magnitude of each of the integrals.[in] rho_0: A density [kg m-3], that is used to calculate the pressure (as p~=-z*rho_0*G_e) used in the equation of state.[in] g_e: The Earth’s gravitational acceleration [m2 Z-1 s-2 ~> m s-2].eos: Equation of state structure[out] dpa: The change in the pressure anomaly[out] intz_dpa: The integral through the thickness of the[out] intx_dpa: The integral in x of the difference between[out] inty_dpa: The integral in y of the difference between[in] bathyt: The depth of the bathymetry [Z ~> m].[in] dz_neglect: A miniscule thickness change [Z ~> m].[in] usemasswghtinterp: If true, uses mass weighting to interpolate T/S for top and bottom integrals.
-
subroutine, public mom_eos::int_density_dz_generic_plm(T_t T_t, T_b T_b, S_t S_t, S_b S_b, z_t z_t, z_b z_b, rho_ref rho_ref, rho_0 rho_0, G_e G_e, dz_subroundoff dz_subroundoff, bathyT bathyT, HII HII, HIO HIO, EOS EOS, dpa dpa, intz_dpa intz_dpa, intx_dpa intx_dpa, inty_dpa inty_dpa, useMassWghtInterp useMassWghtInterp) Compute pressure gradient force integrals by quadrature for the case where T and S are linear profiles.
- Parameters
[in] hii: Ocean horizontal index structures for the input arrays[in] hio: Ocean horizontal index structures for the output arrays[in] t_t: Potential temperatue at the cell top [degC][in] t_b: Potential temperatue at the cell bottom [degC][in] s_t: Salinity at the cell top [ppt][in] s_b: Salinity at the cell bottom [ppt][in] z_t: The geometric height at the top of the layer,[in] z_b: The geometric height at the bottom of the layer [Z ~> m].[in] rho_ref: A mean density [kg m-3], that is subtracted out to reduce the magnitude of each of the integrals.[in] rho_0: A density [kg m-3], that is used to calculate the pressure (as p~=-z*rho_0*G_e) used in the equation of state.[in] g_e: The Earth’s gravitational acceleration [m2 Z-1 s-2 ~> m s-2].[in] dz_subroundoff: A miniscule thickness change [Z ~> m].[in] bathyt: The depth of the bathymetry [Z ~> m].eos: Equation of state structure[out] dpa: The change in the pressure anomaly across the layer [Pa].[out] intz_dpa: The integral through the thickness of the layer of[out] intx_dpa: The integral in x of the difference between the[out] inty_dpa: The integral in y of the difference between the[in] usemasswghtinterp: If true, uses mass weighting to interpolate T/S for top and bottom integrals.
-
subroutine, public mom_eos::find_depth_of_pressure_in_cell(T_t T_t, T_b T_b, S_t S_t, S_b S_b, z_t z_t, z_b z_b, P_t P_t, P_tgt P_tgt, rho_ref rho_ref, G_e G_e, EOS EOS, P_b P_b, z_out z_out, z_tol z_tol) Find the depth at which the reconstructed pressure matches P_tgt.
- Parameters
[in] t_t: Potential temperatue at the cell top [degC][in] t_b: Potential temperatue at the cell bottom [degC][in] s_t: Salinity at the cell top [ppt][in] s_b: Salinity at the cell bottom [ppt][in] z_t: Absolute height of top of cell [Z ~> m]. (Boussinesq ????)[in] z_b: Absolute height of bottom of cell [Z ~> m].[in] p_t: Anomalous pressure of top of cell, relative to g*rho_ref*z_t [Pa][in] p_tgt: Target pressure at height z_out, relative to g*rho_ref*z_out [Pa][in] rho_ref: Reference density with which calculation are anomalous to[in] g_e: Gravitational acceleration [m2 Z-1 s-2 ~> m s-2]eos: Equation of state structure[out] p_b: Pressure at the bottom of the cell [Pa][out] z_out: Absolute depth at which anomalous pressure = p_tgt [Z ~> m].[in] z_tol: The tolerance in finding z_out [Z ~> m].
-
real function mom_eos::frac_dp_at_pos(T_t T_t, T_b T_b, S_t S_t, S_b S_b, z_t z_t, z_b z_b, rho_ref rho_ref, G_e G_e, pos pos, EOS EOS) Returns change in anomalous pressure change from top to non-dimensional position pos between z_t and z_b.
- Parameters
[in] t_t: Potential temperatue at the cell top [degC][in] t_b: Potential temperatue at the cell bottom [degC][in] s_t: Salinity at the cell top [ppt][in] s_b: Salinity at the cell bottom [ppt][in] z_t: The geometric height at the top of the layer [Z ~> m][in] z_b: The geometric height at the bottom of the layer [Z ~> m][in] rho_ref: A mean density [kg m-3], that is subtracted out to reduce the magnitude of each of the integrals.[in] g_e: The Earth’s gravitational acceleration [m s-2][in] pos: The fractional vertical position, 0 to 1 [nondim].eos: Equation of state structure
-
subroutine, public mom_eos::int_density_dz_generic_ppm(T T, T_t T_t, T_b T_b, S S, S_t S_t, S_b S_b, z_t z_t, z_b z_b, rho_ref rho_ref, rho_0 rho_0, G_e G_e, HII HII, HIO HIO, EOS EOS, dpa dpa, intz_dpa intz_dpa, intx_dpa intx_dpa, inty_dpa inty_dpa) Compute pressure gradient force integrals for the case where T and S are parabolic profiles.
- Parameters
[in] hii: Ocean horizontal index structures for the input arrays[in] hio: Ocean horizontal index structures for the output arrays[in] t: Potential temperature referenced to the surface [degC][in] t_t: Potential temperatue at the cell top [degC][in] t_b: Potential temperatue at the cell bottom [degC][in] s: Salinity [ppt][in] s_t: Salinity at the cell top [ppt][in] s_b: Salinity at the cell bottom [ppt][in] z_t: Height at the top of the layer [Z ~> m].[in] z_b: Height at the bottom of the layer [Z ~> m].[in] rho_ref: A mean density [kg m-3], that is subtracted out to reduce the magnitude of each of the integrals.[in] rho_0: A density [kg m-3], that is used to calculate the pressure (as p~=-z*rho_0*G_e) used in the equation of state.[in] g_e: The Earth’s gravitational acceleration [m s-2]eos: Equation of state structure[out] dpa: The change in the pressure anomaly across the layer [Pa].[out] intz_dpa: The integral through the thickness of the layer of[out] intx_dpa: The integral in x of the difference between the[out] inty_dpa: The integral in y of the difference between the
-
subroutine
compute_integral_quadratic(x x, y y, f f, integral integral)¶ Compute the integral of the quadratic function.
- Parameters
[in] x: The x-position of the corners[in] y: The y-position of the corners[in] f: The function at the quadrature points[out] integral: The returned integral
-
subroutine
evaluate_shape_bilinear(xi xi, eta eta, phi phi, dphidxi dphidxi, dphideta dphideta)¶ Evaluation of the four bilinear shape fn and their gradients at (xi,eta)
- Parameters
[in] xi: The x position to evaluate[in] eta: The z position to evaluate[out] phi: The weights of the four corners at this point[out] dphidxi: The x-gradient of the weights of the four corners at this point[out] dphideta: The z-gradient of the weights of the four corners at this point
-
subroutine
evaluate_shape_quadratic(xi xi, eta eta, phi phi, dphidxi dphidxi, dphideta dphideta)¶ Evaluation of the nine quadratic shape fn weights and their gradients at (xi,eta)
- Parameters
[in] xi: The x position to evaluate[in] eta: The z position to evaluate[out] phi: The weights of the 9 bilinear quadrature points at this point[out] dphidxi: The x-gradient of the weights of the 9 bilinear quadrature points corners at this point[out] dphideta: The z-gradient of the weights of the 9 bilinear quadrature points corners at this point
-
subroutine, public mom_eos::int_spec_vol_dp_generic(T T, S S, p_t p_t, p_b p_b, alpha_ref alpha_ref, HI HI, EOS EOS, dza dza, intp_dza intp_dza, intx_dza intx_dza, inty_dza inty_dza, halo_size halo_size, bathyP bathyP, dP_neglect dP_neglect, useMassWghtInterp useMassWghtInterp) This subroutine calculates integrals of specific volume anomalies in pressure across layers, which are required for calculating the finite-volume form pressure accelerations in a non-Boussinesq model. There are essentially no free assumptions, apart from the use of Bode’s rule quadrature to do the integrals.
- Parameters
[in] hi: A horizontal index type structure.[in] t: Potential temperature of the layer [degC].[in] s: Salinity of the layer [ppt].[in] p_t: Pressure atop the layer [Pa].[in] p_b: Pressure below the layer [Pa].[in] alpha_ref: A mean specific volume that is subtracted out to reduce the magnitude of each of the integrals [m3 kg-1]. The calculation is mathematically identical with different values of alpha_ref, but alpha_ref alters the effects of roundoff, and answers do change.eos: Equation of state structure[out] dza: The change in the geopotential anomaly[out] intp_dza: The integral in pressure through the[out] intx_dza: The integral in x of the difference[out] inty_dza: The integral in y of the difference[in] halo_size: The width of halo points on which to calculate dza.[in] bathyp: The pressure at the bathymetry [Pa][in] dp_neglect: A miniscule pressure change with the same units as p_t (Pa?)[in] usemasswghtinterp: If true, uses mass weighting to interpolate T/S for top and bottom integrals.
-
subroutine, public mom_eos::int_spec_vol_dp_generic_plm(T_t T_t, T_b T_b, S_t S_t, S_b S_b, p_t p_t, p_b p_b, alpha_ref alpha_ref, dP_neglect dP_neglect, bathyP bathyP, HI HI, EOS EOS, dza dza, intp_dza intp_dza, intx_dza intx_dza, inty_dza inty_dza, useMassWghtInterp useMassWghtInterp) This subroutine calculates integrals of specific volume anomalies in pressure across layers, which are required for calculating the finite-volume form pressure accelerations in a non-Boussinesq model. There are essentially no free assumptions, apart from the use of Bode’s rule quadrature to do the integrals.
- Parameters
[in] hi: A horizontal index type structure.[in] t_t: Potential temperature at the top of the layer [degC].[in] t_b: Potential temperature at the bottom of the layer [degC].[in] s_t: Salinity at the top the layer [ppt].[in] s_b: Salinity at the bottom the layer [ppt].[in] p_t: Pressure atop the layer [Pa].[in] p_b: Pressure below the layer [Pa].[in] alpha_ref: A mean specific volume that is subtracted out to reduce the magnitude of each of the integrals [m3 kg-1]. The calculation is mathematically identical with different values of alpha_ref, but alpha_ref alters the effects of roundoff, and answers do change.[in] dp_neglect: A miniscule pressure change with the same units as p_t (Pa?)[in] bathyp: The pressure at the bathymetry [Pa]eos: Equation of state structure[out] dza: The change in the geopotential anomaly[out] intp_dza: The integral in pressure through the[out] intx_dza: The integral in x of the difference[out] inty_dza: The integral in y of the difference[in] usemasswghtinterp: If true, uses mass weighting to interpolate T/S for top and bottom integrals.
-
subroutine, public mom_eos::convert_temp_salt_for_teos10(T T, S S, press press, G G, kd kd, mask_z mask_z, EOS EOS) Convert T&S to Absolute Salinity and Conservative Temperature if using TEOS10.
- Parameters
[in] g: The ocean’s grid structure[inout] t: Potential temperature referenced to the surface [degC][inout] s: Salinity [ppt][in] press: Pressure at the top of the layer [Pa].eos: Equation of state structure[in] mask_z: 3d mask regulating which points to convert.[in] kd: The number of layers to work on
-
subroutine, public mom_eos::extract_member_eos(EOS EOS, form_of_EOS form_of_EOS, form_of_TFreeze form_of_TFreeze, EOS_quadrature EOS_quadrature, Compressible Compressible, Rho_T0_S0 Rho_T0_S0, drho_dT drho_dT, dRho_dS dRho_dS, TFr_S0_P0 TFr_S0_P0, dTFr_dS dTFr_dS, dTFr_dp dTFr_dp) Extractor routine for the EOS type if the members need to be accessed outside this module.
- Parameters
eos: Equation of state structure[out] form_of_eos: A coded integer indicating the equation of state to use.[out] form_of_tfreeze: A coded integer indicating the expression for the potential temperature of the freezing point.[out] eos_quadrature: If true, always use the generic (quadrature) code for the integrals of density.[out] compressible: If true, in situ density is a function of pressure.[out] rho_t0_s0: Density at T=0 degC and S=0 ppt [kg m-3][out] drho_dt: Partial derivative of density with temperature in [kg m-3 degC-1][out] drho_ds: Partial derivative of density with salinity in [kg m-3 ppt-1][out] tfr_s0_p0: The freezing potential temperature at S=0, P=0 [degC].[out] dtfr_ds: The derivative of freezing point with salinity [degC PSU-1].[out] dtfr_dp: The derivative of freezing point with pressure [degC Pa-1].
Variables
-
integer, parameter, public mom_eos::eos_linear = 1 A named integer specifying an equation of state.
-
integer, parameter, public mom_eos::eos_unesco = 2 A named integer specifying an equation of state.
-
integer, parameter, public mom_eos::eos_wright = 3 A named integer specifying an equation of state.
-
integer, parameter, public mom_eos::eos_teos10 = 4 A named integer specifying an equation of state.
-
integer, parameter, public mom_eos::eos_nemo = 5 A named integer specifying an equation of state.
-
character*(10), parameter mom_eos::eos_linear_string = "LINEAR" A string for specifying the equation of state.
-
character*(10), parameter mom_eos::eos_unesco_string = "UNESCO" A string for specifying the equation of state.
-
character*(10), parameter mom_eos::eos_wright_string = "WRIGHT" A string for specifying the equation of state.
-
character*(10), parameter mom_eos::eos_teos10_string = "TEOS10" A string for specifying the equation of state.
-
character*(10), parameter mom_eos::eos_nemo_string = "NEMO" A string for specifying the equation of state.
-
character*(10), parameter mom_eos::eos_default = EOS_WRIGHT_STRING The default equation of state.
-
integer, parameter mom_eos::tfreeze_linear = 1 A named integer specifying a freezing point expression.
-
integer, parameter mom_eos::tfreeze_millero = 2 A named integer specifying a freezing point expression.
-
integer, parameter mom_eos::tfreeze_teos10 = 3 A named integer specifying a freezing point expression.
-
character*(10), parameter mom_eos::tfreeze_linear_string = "LINEAR" A string for specifying the freezing point expression.
-
character*(10), parameter mom_eos::tfreeze_millero_string = "MILLERO_78" A string for specifying freezing point expression.
-
character*(10), parameter mom_eos::tfreeze_teos10_string = "TEOS10" A string for specifying the freezing point expression.
-
character*(10), parameter mom_eos::tfreeze_default = TFREEZE_LINEAR_STRING The default freezing point expression.
-
subroutine
-
namespace
mom_eos_linear¶ A simple linear equation of state for sea water with constant coefficients.
Functions
-
subroutine, public mom_eos_linear::calculate_density_scalar_linear(T T, S S, pressure pressure, rho rho, Rho_T0_S0 Rho_T0_S0, dRho_dT dRho_dT, dRho_dS dRho_dS, rho_ref rho_ref) This subroutine computes the density of sea water with a trivial linear equation of state (in [kg m-3]) from salinity (sal [PSU]), potential temperature (T [degC]), and pressure [Pa].
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: pressure [Pa].[out] rho: In situ density [kg m-3].[in] rho_t0_s0: The density at T=0, S=0 [kg m-3].[in] drho_dt: The derivatives of density with temperature [kg m-3 degC-1].[in] drho_ds: The derivatives of density with salinity in [kg m-3 ppt-1].[in] rho_ref: A reference density [kg m-3].
-
subroutine, public mom_eos_linear::calculate_density_array_linear(T T, S S, pressure pressure, rho rho, start start, npts npts, Rho_T0_S0 Rho_T0_S0, dRho_dT dRho_dT, dRho_dS dRho_dS, rho_ref rho_ref) This subroutine computes the density of sea water with a trivial linear equation of state (in kg/m^3) from salinity (sal in psu), potential temperature (T [degC]), and pressure [Pa].
- Parameters
[in] t: potential temperature relative to the surface [degC].[in] s: salinity [PSU].[in] pressure: pressure [Pa].[out] rho: in situ density [kg m-3].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.[in] rho_t0_s0: The density at T=0, S=0 [kg m-3].[in] drho_dt: The derivatives of density with temperature [kg m-3 degC-1].[in] drho_ds: The derivatives of density with salinity in [kg m-3 ppt-1].[in] rho_ref: A reference density [kg m-3].
-
subroutine
calculate_spec_vol_scalar_linear(T T, S S, pressure pressure, specvol specvol, Rho_T0_S0 Rho_T0_S0, dRho_dT dRho_dT, dRho_dS dRho_dS, spv_ref spv_ref)¶ This subroutine computes the in situ specific volume of sea water (specvol in [m3 kg-1]) from salinity (S [PSU]), potential temperature (T [degC]) and pressure [Pa], using a trivial linear equation of state for density. If spv_ref is present, specvol is an anomaly from spv_ref.
- Parameters
[in] t: potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: Pressure [Pa].[out] specvol: In situ specific volume [m3 kg-1].[in] rho_t0_s0: The density at T=0, S=0 [kg m-3].[in] drho_dt: The derivatives of density with temperature [kg m-3 degC-1].[in] drho_ds: The derivatives of density with salinity [kg m-3 ppt-1].[in] spv_ref: A reference specific volume [m3 kg-1].
-
subroutine
calculate_spec_vol_array_linear(T T, S S, pressure pressure, specvol specvol, start start, npts npts, Rho_T0_S0 Rho_T0_S0, dRho_dT dRho_dT, dRho_dS dRho_dS, spv_ref spv_ref)¶ This subroutine computes the in situ specific volume of sea water (specvol in [m3 kg-1]) from salinity (S [PSU]), potential temperature (T [degC]) and pressure [Pa], using a trivial linear equation of state for density. If spv_ref is present, specvol is an anomaly from spv_ref.
- Parameters
[in] t: potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: Pressure [Pa].[out] specvol: in situ specific volume [m3 kg-1].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.[in] rho_t0_s0: The density at T=0, S=0 [kg m-3].[in] drho_dt: The derivatives of density with temperature [kg m-3 degC-1].[in] drho_ds: The derivatives of density with salinity [kg m-3 ppt-1].[in] spv_ref: A reference specific volume [m3 kg-1].
-
subroutine
calculate_density_derivs_array_linear(T T, S S, pressure pressure, drho_dT_out drho_dT_out, drho_dS_out drho_dS_out, Rho_T0_S0 Rho_T0_S0, dRho_dT dRho_dT, dRho_dS dRho_dS, start start, npts npts)¶ This subroutine calculates the partial derivatives of density * with potential temperature and salinity.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: Pressure [Pa].[out] drho_dt_out: The partial derivative of density with potential temperature [kg m-3 degC-1].[out] drho_ds_out: The partial derivative of density with salinity [kg m-3 ppt-1].[in] rho_t0_s0: The density at T=0, S=0 [kg m-3].[in] drho_dt: The derivative of density with temperature [kg m-3 degC-1].[in] drho_ds: The derivative of density with salinity [kg m-3 ppt-1].[in] start: The starting point in the arrays.[in] npts: The number of values to calculate.
-
subroutine, public mom_eos_linear::calculate_density_derivs_scalar_linear(T T, S S, pressure pressure, drho_dT_out drho_dT_out, drho_dS_out drho_dS_out, Rho_T0_S0 Rho_T0_S0, dRho_dT dRho_dT, dRho_dS dRho_dS) This subroutine calculates the partial derivatives of density * with potential temperature and salinity for a single point.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: pressure [Pa].[out] drho_dt_out: The partial derivative of density with potential temperature [kg m-3 degC-1].[out] drho_ds_out: The partial derivative of density with salinity [kg m-3 ppt-1].[in] rho_t0_s0: The density at T=0, S=0 [kg m-3].[in] drho_dt: The derivatives of density with temperature [kg m-3 degC-1].[in] drho_ds: The derivatives of density with salinity [kg m-3 ppt-1].
-
subroutine
calculate_density_second_derivs_scalar_linear(T T, S S, pressure pressure, drho_dS_dS drho_dS_dS, drho_dS_dT drho_dS_dT, drho_dT_dT drho_dT_dT, drho_dS_dP drho_dS_dP, drho_dT_dP drho_dT_dP)¶ This subroutine calculates the five, partial second derivatives of density w.r.t. potential temperature and salinity and pressure which for a linear equation of state should all be 0.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: pressure [Pa].[out] drho_ds_ds: The second derivative of density with salinity [kg m-3 PSU-2].[out] drho_ds_dt: The second derivative of density with temperature and salinity [kg m-3 ppt-1 degC-1].[out] drho_dt_dt: The second derivative of density with temperature [kg m-3 degC-2].[out] drho_ds_dp: The second derivative of density with salinity and pressure [kg m-3 PSU-1 Pa-1].[out] drho_dt_dp: The second derivative of density with temperature and pressure [kg m-3 degC-1 Pa-1].
-
subroutine
calculate_density_second_derivs_array_linear(T T, S S, pressure pressure, drho_dS_dS drho_dS_dS, drho_dS_dT drho_dS_dT, drho_dT_dT drho_dT_dT, drho_dS_dP drho_dS_dP, drho_dT_dP drho_dT_dP, start start, npts npts)¶ This subroutine calculates the five, partial second derivatives of density w.r.t. potential temperature and salinity and pressure which for a linear equation of state should all be 0.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: pressure [Pa].[out] drho_ds_ds: The second derivative of density with salinity [kg m-3 PSU-2].[out] drho_ds_dt: The second derivative of density with temperature and salinity [kg m-3 ppt-1 degC-1].[out] drho_dt_dt: The second derivative of density with temperature [kg m-3 degC-2].[out] drho_ds_dp: The second derivative of density with salinity and pressure [kg m-3 PSU-1 Pa-1].[out] drho_dt_dp: The second derivative of density with temperature and pressure [kg m-3 degC-1 Pa-1].[in] start: The starting point in the arrays.[in] npts: The number of values to calculate.
-
subroutine, public mom_eos_linear::calculate_specvol_derivs_linear(T T, S S, pressure pressure, dSV_dT dSV_dT, dSV_dS dSV_dS, start start, npts npts, Rho_T0_S0 Rho_T0_S0, dRho_dT dRho_dT, dRho_dS dRho_dS) Calculate the derivatives of specific volume with temperature and salinity.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: pressure [Pa].[out] dsv_ds: The partial derivative of specific volume with salinity [m3 kg-1 PSU-1].[out] dsv_dt: The partial derivative of specific volume with potential temperature [m3 kg-1 degC-1].[in] start: The starting point in the arrays.[in] npts: The number of values to calculate.[in] rho_t0_s0: The density at T=0, S=0 [kg m-3].[in] drho_dt: The derivative of density with temperature, [kg m-3 degC-1].[in] drho_ds: The derivative of density with salinity [kg m-3 ppt-1].
-
subroutine, public mom_eos_linear::calculate_compress_linear(T T, S S, pressure pressure, rho rho, drho_dp drho_dp, start start, npts npts, Rho_T0_S0 Rho_T0_S0, dRho_dT dRho_dT, dRho_dS dRho_dS) This subroutine computes the in situ density of sea water (rho) and the compressibility (drho/dp == C_sound^-2) at the given salinity, potential temperature, and pressure.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: pressure [Pa].[out] rho: In situ density [kg m-3].[out] drho_dp: The partial derivative of density with pressure (also the inverse of the square of sound speed) [s2 m-2].[in] start: The starting point in the arrays.[in] npts: The number of values to calculate.[in] rho_t0_s0: The density at T=0, S=0 [kg m-3].[in] drho_dt: The derivative of density with temperature [kg m-3 degC-1].[in] drho_ds: The derivative of density with salinity [kg m-3 ppt-1].
-
subroutine, public mom_eos_linear::int_density_dz_linear(T T, S S, z_t z_t, z_b z_b, rho_ref rho_ref, rho_0_pres rho_0_pres, G_e G_e, HII HII, HIO HIO, Rho_T0_S0 Rho_T0_S0, dRho_dT dRho_dT, dRho_dS dRho_dS, dpa dpa, intz_dpa intz_dpa, intx_dpa intx_dpa, inty_dpa inty_dpa, bathyT bathyT, dz_neglect dz_neglect, useMassWghtInterp useMassWghtInterp) This subroutine calculates analytical and nearly-analytical integrals of pressure anomalies across layers, which are required for calculating the finite-volume form pressure accelerations in a Boussinesq model.
- Parameters
[in] hii: The horizontal index type for the input arrays.[in] hio: The horizontal index type for the output arrays.[in] t: Potential temperature relative to the surface[in] s: Salinity [PSU].[in] z_t: Height at the top of the layer in depth units [Z ~> m].[in] z_b: Height at the top of the layer [Z ~> m].[in] rho_ref: A mean density [kg m-3], that is subtracted out to reduce the magnitude of each of the integrals.[in] rho_0_pres: A density [kg m-3], that is used to calculate the pressure (as p~=-z*rho_0_pres*G_e) used in the equation of state. rho_0_pres is not used here.[in] g_e: The Earth’s gravitational acceleration [m2 Z-1 s-2 ~> m s-2].[in] rho_t0_s0: The density at T=0, S=0 [kg m-3].[in] drho_dt: The derivative of density with temperature, [kg m-3 degC-1].[in] drho_ds: The derivative of density with salinity, in [kg m-3 ppt-1].[out] dpa: The change in the pressure anomaly across the[out] intz_dpa: The integral through the thickness of the layer[out] intx_dpa: The integral in x of the difference between the[out] inty_dpa: The integral in y of the difference between the[in] bathyt: The depth of the bathymetry [Z ~> m].[in] dz_neglect: A miniscule thickness change [Z ~> m].[in] usemasswghtinterp: If true, uses mass weighting to interpolate T/S for top and bottom integrals.
-
subroutine, public mom_eos_linear::int_spec_vol_dp_linear(T T, S S, p_t p_t, p_b p_b, alpha_ref alpha_ref, HI HI, Rho_T0_S0 Rho_T0_S0, dRho_dT dRho_dT, dRho_dS dRho_dS, dza dza, intp_dza intp_dza, intx_dza intx_dza, inty_dza inty_dza, halo_size halo_size, bathyP bathyP, dP_neglect dP_neglect, useMassWghtInterp useMassWghtInterp) Calculates analytical and nearly-analytical integrals in pressure across layers of geopotential anomalies, which are required for calculating the finite-volume form pressure accelerations in a non-Boussinesq model. Specific volume is assumed to vary linearly between adjacent points.
- Parameters
[in] hi: The ocean’s horizontal index type.[in] t: Potential temperature relative to the surface[in] s: Salinity [PSU].[in] p_t: Pressure at the top of the layer [Pa].[in] p_b: Pressure at the top of the layer [Pa].[in] alpha_ref: A mean specific volume that is subtracted out to reduce the magnitude of each of the integrals, m3 kg-1. The calculation is mathematically identical with different values of alpha_ref, but this reduces the effects of roundoff.[in] rho_t0_s0: The density at T=0, S=0 [kg m-3].[in] drho_dt: The derivative of density with temperature [kg m-3 degC-1].[in] drho_ds: The derivative of density with salinity, in [kg m-3 ppt-1].[out] dza: The change in the geopotential anomaly across[out] intp_dza: The integral in pressure through the layer of[out] intx_dza: The integral in x of the difference between the[out] inty_dza: The integral in y of the difference between the[in] halo_size: The width of halo points on which to calculate dza.[in] bathyp: The pressure at the bathymetry [Pa][in] dp_neglect: A miniscule pressure change with the same units as p_t [Pa][in] usemasswghtinterp: If true, uses mass weighting to interpolate T/S for top and bottom integrals.
-
-
namespace
mom_eos_nemo¶ The equation of state using the expressions of Roquet et al. that are used in NEMO.
Unnamed Group
-
real, parameter mom_eos_nemo::rdeltas = 32. Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::r1_s0 = 0.875/35.16504 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::r1_t0 = 1./40. Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::r1_p0 = 1.e-4 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::r00 = 4.6494977072e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::r01 = -5.2099962525 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::r02 = 2.2601900708e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::r03 = 6.4326772569e-02 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::r04 = 1.5616995503e-02 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::r05 = -1.7243708991e-03 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos000 = 8.0189615746e+02 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos100 = 8.6672408165e+02 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos200 = -1.7864682637e+03 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos300 = 2.0375295546e+03 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos400 = -1.2849161071e+03 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos500 = 4.3227585684e+02 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos600 = -6.0579916612e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos010 = 2.6010145068e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos110 = -6.5281885265e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos210 = 8.1770425108e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos310 = -5.6888046321e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos410 = 1.7681814114e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos510 = -1.9193502195 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos020 = -3.7074170417e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos120 = 6.1548258127e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos220 = -6.0362551501e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos320 = 2.9130021253e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos420 = -5.4723692739 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos030 = 2.1661789529e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos130 = -3.3449108469e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos230 = 1.9717078466e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos330 = -3.1742946532 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos040 = -8.3627885467 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos140 = 1.1311538584e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos240 = -5.3563304045 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos050 = 5.4048723791e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos150 = 4.8169980163e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos060 = -1.9083568888e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos001 = 1.9681925209e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos101 = -4.2549998214e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos201 = 5.0774768218e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos301 = -3.0938076334e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos401 = 6.6051753097 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos011 = -1.3336301113e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos111 = -4.4870114575 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos211 = 5.0042598061 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos311 = -6.5399043664e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos021 = 6.7080479603 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos121 = 3.5063081279 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos221 = -1.8795372996 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos031 = -2.4649669534 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos131 = -5.5077101279e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos041 = 5.5927935970e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos002 = 2.0660924175 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos102 = -4.9527603989 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos202 = 2.5019633244 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos012 = 2.0564311499 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos112 = -2.1311365518e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos022 = -1.2419983026 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos003 = -2.3342758797e-02 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos103 = -1.8507636718e-02 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::eos013 = 3.7969820455e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp000 = -6.5025362670e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp100 = 1.6320471316 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp200 = -2.0442606277 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp300 = 1.4222011580 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp400 = -4.4204535284e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp500 = 4.7983755487e-02 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp010 = 1.8537085209 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp110 = -3.0774129064 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp210 = 3.0181275751 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp310 = -1.4565010626 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp410 = 2.7361846370e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp020 = -1.6246342147 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp120 = 2.5086831352 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp220 = -1.4787808849 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp320 = 2.3807209899e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp030 = 8.3627885467e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp130 = -1.1311538584 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp230 = 5.3563304045e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp040 = -6.7560904739e-02 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp140 = -6.0212475204e-02 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp050 = 2.8625353333e-02 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp001 = 3.3340752782e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp101 = 1.1217528644e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp201 = -1.2510649515e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp301 = 1.6349760916e-02 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp011 = -3.3540239802e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp111 = -1.7531540640e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp211 = 9.3976864981e-02 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp021 = 1.8487252150e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp121 = 4.1307825959e-02 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp031 = -5.5927935970e-02 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp002 = -5.1410778748e-02 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp102 = 5.3278413794e-03 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp012 = 6.2099915132e-02 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::alp003 = -9.4924551138e-03 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet000 = 1.0783203594e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet100 = -4.4452095908e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet200 = 7.6048755820e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet300 = -6.3944280668e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet400 = 2.6890441098e+01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet500 = -4.5221697773 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet010 = -8.1219372432e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet110 = 2.0346663041 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet210 = -2.1232895170 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet310 = 8.7994140485e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet410 = -1.1939638360e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet020 = 7.6574242289e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet120 = -1.5019813020 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet220 = 1.0872489522 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet320 = -2.7233429080e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet030 = -4.1615152308e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet130 = 4.9061350869e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet230 = -1.1847737788e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet040 = 1.4073062708e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet140 = -1.3327978879e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet050 = 5.9929880134e-03 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet001 = -5.2937873009e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet101 = 1.2634116779 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet201 = -1.1547328025 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet301 = 3.2870876279e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet011 = -5.5824407214e-02 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet111 = 1.2451933313e-01 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet211 = -2.4409539932e-02 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet021 = 4.3623149752e-02 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet121 = -4.6767901790e-02 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet031 = -6.8523260060e-03 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet002 = -6.1618945251e-02 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet102 = 6.2255521644e-02 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet012 = -2.6514181169e-03 Parameters in the NEMO equation of state.
-
real, parameter mom_eos_nemo::bet003 = -2.3025968587e-04 Parameters in the NEMO equation of state.
-
subroutine, public mom_eos_nemo::calculate_density_scalar_nemo(T T, S S, pressure pressure, rho rho, rho_ref rho_ref) This subroutine computes the in situ density of sea water (rho in [kg m-3]) from absolute salinity (S [g kg-1]), conservative temperature (T [degC]), and pressure [Pa]. It uses the expressions derived for use with NEMO.
- Parameters
[in] t: Conservative temperature [degC].[in] s: Absolute salinity [g kg-1].[in] pressure: pressure [Pa].[out] rho: In situ density [kg m-3].[in] rho_ref: A reference density [kg m-3].
-
subroutine, public mom_eos_nemo::calculate_density_array_nemo(T T, S S, pressure pressure, rho rho, start start, npts npts, rho_ref rho_ref) This subroutine computes the in situ density of sea water (rho in [kg m-3]) from absolute salinity (S [g kg-1]), conservative temperature (T [degC]), and pressure [Pa]. It uses the expressions derived for use with NEMO.
- Parameters
[in] t: Conservative temperature [degC].[in] s: Absolute salinity [g kg-1].[in] pressure: pressure [Pa].[out] rho: in situ density [kg m-3].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.[in] rho_ref: A reference density [kg m-3].
-
subroutine
calculate_density_derivs_array_nemo(T T, S S, pressure pressure, drho_dT drho_dT, drho_dS drho_dS, start start, npts npts)¶ For a given thermodynamic state, calculate the derivatives of density with conservative temperature and absolute salinity, using the expressions derived for use with NEMO.
- Parameters
[in] t: Conservative temperature [degC].[in] s: Absolute salinity [g kg-1].[in] pressure: pressure [Pa].[out] drho_dt: The partial derivative of density with potential temperature [kg m-3 degC-1].[out] drho_ds: The partial derivative of density with salinity, in [kg m-3 ppt-1].[in] start: The starting point in the arrays.[in] npts: The number of values to calculate.
-
subroutine
calculate_density_derivs_scalar_nemo(T T, S S, pressure pressure, drho_dt drho_dt, drho_ds drho_ds)¶ Wrapper to calculate_density_derivs_array for scalar inputs.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [g kg-1].[in] pressure: Pressure [Pa].[out] drho_dt: The partial derivative of density with potential temperature [kg m-3 degC-1].[out] drho_ds: The partial derivative of density with salinity, in [kg m-3 ppt-1].
-
subroutine, public mom_eos_nemo::calculate_compress_nemo(T T, S S, pressure pressure, rho rho, drho_dp drho_dp, start start, npts npts) Compute the in situ density of sea water (rho in [kg m-3]) and the compressibility (drho/dp = C_sound^-2, stored as drho_dp [s2 m-2]) from absolute salinity (sal in g/kg), conservative temperature (T [degC]), and pressure [Pa], using the expressions derived for use with NEMO.
- Parameters
[in] t: Conservative temperature [degC].[in] s: Absolute salinity [g/kg].[in] pressure: pressure [Pa].[out] rho: In situ density [kg m-3].[out] drho_dp: The partial derivative of density with pressure (also the inverse of the square of sound speed) [s2 m-2].[in] start: The starting point in the arrays.[in] npts: The number of values to calculate.
Variables
-
real, parameter mom_eos_nemo::pa2db = 1.e-4 Conversion factor between Pa and dbar.
-
-
namespace
mom_eos_teos10¶ The equation of state using the TEOS10 expressions.
Functions
-
subroutine
calculate_density_scalar_teos10(T T, S S, pressure pressure, rho rho, rho_ref rho_ref)¶ This subroutine computes the in situ density of sea water (rho in [kg m-3]) from absolute salinity (S [g kg-1]), conservative temperature (T [degC]), and pressure [Pa]. It uses the expression from the TEOS10 website.
- Parameters
[in] t: Conservative temperature [degC].[in] s: Absolute salinity [g kg-1].[in] pressure: pressure [Pa].[out] rho: In situ density [kg m-3].[in] rho_ref: A reference density [kg m-3].
-
subroutine
calculate_density_array_teos10(T T, S S, pressure pressure, rho rho, start start, npts npts, rho_ref rho_ref)¶ This subroutine computes the in situ density of sea water (rho in [kg m-3]) from absolute salinity (S [g kg-1]), conservative temperature (T [degC]), and pressure [Pa]. It uses the expression from the TEOS10 website.
- Parameters
[in] t: Conservative temperature [degC].[in] s: Absolute salinity [g kg-1][in] pressure: pressure [Pa].[out] rho: in situ density [kg m-3].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.[in] rho_ref: A reference density [kg m-3].
-
subroutine
calculate_spec_vol_scalar_teos10(T T, S S, pressure pressure, specvol specvol, spv_ref spv_ref)¶ This subroutine computes the in situ specific volume of sea water (specvol in [m3 kg-1]) from absolute salinity (S [g kg-1]), conservative temperature (T [degC]) and pressure [Pa], using the TEOS10 equation of state. If spv_ref is present, specvol is an anomaly from spv_ref.
- Parameters
[in] t: Conservative temperature [degC].[in] s: Absolute salinity [g kg-1][in] pressure: pressure [Pa].[out] specvol: in situ specific volume [m3 kg-1].[in] spv_ref: A reference specific volume [m3 kg-1].
-
subroutine
calculate_spec_vol_array_teos10(T T, S S, pressure pressure, specvol specvol, start start, npts npts, spv_ref spv_ref)¶ This subroutine computes the in situ specific volume of sea water (specvol in [m3 kg-1]) from absolute salinity (S [g kg-1]), conservative temperature (T [degC]) and pressure [Pa], using the TEOS10 equation of state. If spv_ref is present, specvol is an anomaly from spv_ref.
- Parameters
[in] t: Conservative temperature relative to the surface [degC].[in] s: salinity [g kg-1].[in] pressure: pressure [Pa].[out] specvol: in situ specific volume [m3 kg-1].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.[in] spv_ref: A reference specific volume [m3 kg-1].
-
subroutine
calculate_density_derivs_array_teos10(T T, S S, pressure pressure, drho_dT drho_dT, drho_dS drho_dS, start start, npts npts)¶ For a given thermodynamic state, calculate the derivatives of density with conservative temperature and absolute salinity, using the TEOS10 expressions.
- Parameters
[in] t: Conservative temperature [degC].[in] s: Absolute salinity [g kg-1].[in] pressure: pressure [Pa].[out] drho_dt: The partial derivative of density with conservative temperature [kg m-3 degC-1].[out] drho_ds: The partial derivative of density with absolute salinity, [kg m-3 (g/kg)-1].[in] start: The starting point in the arrays.[in] npts: The number of values to calculate.
-
subroutine
calculate_density_derivs_scalar_teos10(T T, S S, pressure pressure, drho_dT drho_dT, drho_dS drho_dS)¶ For a given thermodynamic state, calculate the derivatives of density with conservative temperature and absolute salinity, using the TEOS10 expressions.
- Parameters
[in] t: Conservative temperature [degC][in] s: Absolute Salinity [g kg-1][in] pressure: pressure [Pa].[out] drho_dt: The partial derivative of density with conservative temperature [kg m-3 degC-1].[out] drho_ds: The partial derivative of density with absolute salinity, [kg m-3 (g/kg)-1].
-
subroutine, public mom_eos_teos10::calculate_specvol_derivs_teos10(T T, S S, pressure pressure, dSV_dT dSV_dT, dSV_dS dSV_dS, start start, npts npts) For a given thermodynamic state, calculate the derivatives of specific volume with conservative temperature and absolute salinity, using the TEOS10 expressions.
- Parameters
[in] t: Conservative temperature [degC].[in] s: Absolute salinity [g kg-1].[in] pressure: pressure [Pa].[out] dsv_dt: The partial derivative of specific volume with conservative temperature [m3 kg-1 degC-1].[out] dsv_ds: The partial derivative of specific volume with absolute salinity [m3 kg-1 (g/kg)-1].[in] start: The starting point in the arrays.[in] npts: The number of values to calculate.
-
subroutine
calculate_density_second_derivs_scalar_teos10(T T, S S, pressure pressure, drho_dS_dS drho_dS_dS, drho_dS_dT drho_dS_dT, drho_dT_dT drho_dT_dT, drho_dS_dP drho_dS_dP, drho_dT_dP drho_dT_dP)¶ Calculate the 5 second derivatives of the equation of state for scalar inputs.
- Parameters
[in] t: Conservative temperature [degC][in] s: Absolute Salinity [g kg-1][in] pressure: pressure [Pa].[out] drho_ds_ds: Partial derivative of beta with respect to S[out] drho_ds_dt: Partial derivative of beta with resepct to T[out] drho_dt_dt: Partial derivative of alpha with respect to T[out] drho_ds_dp: Partial derivative of beta with respect to pressure[out] drho_dt_dp: Partial derivative of alpha with respect to pressure
-
subroutine
calculate_density_second_derivs_array_teos10(T T, S S, pressure pressure, drho_dS_dS drho_dS_dS, drho_dS_dT drho_dS_dT, drho_dT_dT drho_dT_dT, drho_dS_dP drho_dS_dP, drho_dT_dP drho_dT_dP, start start, npts npts)¶ Calculate the 5 second derivatives of the equation of state for scalar inputs.
- Parameters
[in] t: Conservative temperature [degC][in] s: Absolute Salinity [g kg-1][in] pressure: pressure [Pa].[out] drho_ds_ds: Partial derivative of beta with respect to S[out] drho_ds_dt: Partial derivative of beta with resepct to T[out] drho_dt_dt: Partial derivative of alpha with respect to T[out] drho_ds_dp: Partial derivative of beta with respect to pressure[out] drho_dt_dp: Partial derivative of alpha with respect to pressure[in] start: The starting point in the arrays.[in] npts: The number of values to calculate.
-
subroutine, public mom_eos_teos10::calculate_compress_teos10(T T, S S, pressure pressure, rho rho, drho_dp drho_dp, start start, npts npts) This subroutine computes the in situ density of sea water (rho in [kg m-3]) and the compressibility (drho/dp = C_sound^-2) (drho_dp [s2 m-2]) from absolute salinity (sal in g/kg), conservative temperature (T [degC]), and pressure [Pa]. It uses the subroutines from TEOS10 website.
- Parameters
[in] t: Conservative temperature [degC].[in] s: Absolute salinity [g kg-1].[in] pressure: Pressure [Pa].[out] rho: In situ density [kg m-3].[out] drho_dp: The partial derivative of density with pressure (also the inverse of the square of sound speed) [s2 m-2].[in] start: The starting point in the arrays.[in] npts: The number of values to calculate.
Variables
-
real, parameter mom_eos_teos10::pa2db = 1.e-4 The conversion factor from Pa to dbar.
-
subroutine
-
namespace
mom_eos_unesco¶ The equation of state using the Jackett and McDougall fits to the UNESCO EOS.
Unnamed Group
-
real, parameter mom_eos_unesco::r00 = 999.842594 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::r10 = 6.793952e-2 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::r20 = -9.095290e-3 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::r30 = 1.001685e-4 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::r40 = -1.120083e-6 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::r50 = 6.536332e-9 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::r01 = 0.824493 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::r11 = -4.0899e-3 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::r21 = 7.6438e-5 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::r31 = -8.2467e-7 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::r41 = 5.3875e-9 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::r032 = -5.72466e-3 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::r132 = 1.0227e-4 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::r232 = -1.6546e-6 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::r02 = 4.8314e-4 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::s00 = 1.965933e4 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::s10 = 1.444304e2 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::s20 = -1.706103 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::s30 = 9.648704e-3 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::s40 = -4.190253e-5 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::s01 = 52.84855 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::s11 = -3.101089e-1 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::s21 = 6.283263e-3 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::s31 = -5.084188e-5 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::s032 = 3.886640e-1 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::s132 = 9.085835e-3 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::s232 = -4.619924e-4 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::sp00 = 3.186519 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::sp10 = 2.212276e-2 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::sp20 = -2.984642e-4 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::sp30 = 1.956415e-6 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::sp01 = 6.704388e-3 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::sp11 = -1.847318e-4 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::sp21 = 2.059331e-7 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::sp032 = 1.480266e-4 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::sp000 = 2.102898e-4 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::sp010 = -1.202016e-5 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::sp020 = 1.394680e-7 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::sp001 = -2.040237e-6 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::sp011 = 6.128773e-8 Parameters in the UNESCO equation of state.
-
real, parameter mom_eos_unesco::sp021 = 6.207323e-10 Parameters in the UNESCO equation of state.
-
subroutine, public mom_eos_unesco::calculate_density_scalar_unesco(T T, S S, pressure pressure, rho rho, rho_ref rho_ref) This subroutine computes the in situ density of sea water (rho in [kg m-3]) from salinity (S [PSU]), potential temperature (T [degC]), and pressure [Pa], using the UNESCO (1981) equation of state.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: pressure [Pa].[out] rho: In situ density [kg m-3].[in] rho_ref: A reference density [kg m-3].
-
subroutine, public mom_eos_unesco::calculate_density_array_unesco(T T, S S, pressure pressure, rho rho, start start, npts npts, rho_ref rho_ref) This subroutine computes the in situ density of sea water (rho in [kg m-3]) from salinity (S [PSU]), potential temperature (T [degC]), and pressure [Pa], using the UNESCO (1981) equation of state.
- Parameters
[in] t: potential temperature relative to the surface [degC].[in] s: salinity [PSU].[in] pressure: pressure [Pa].[out] rho: in situ density [kg m-3].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.[in] rho_ref: A reference density [kg m-3].
-
subroutine
calculate_spec_vol_scalar_unesco(T T, S S, pressure pressure, specvol specvol, spv_ref spv_ref)¶ This subroutine computes the in situ specific volume of sea water (specvol in [m3 kg-1]) from salinity (S [PSU]), potential temperature (T [degC]) and pressure [Pa], using the UNESCO (1981) equation of state. If spv_ref is present, specvol is an anomaly from spv_ref.
- Parameters
[in] t: potential temperature relative to the surface [degC].[in] s: salinity [PSU].[in] pressure: pressure [Pa].[out] specvol: in situ specific volume [m3 kg-1].[in] spv_ref: A reference specific volume [m3 kg-1].
-
subroutine
calculate_spec_vol_array_unesco(T T, S S, pressure pressure, specvol specvol, start start, npts npts, spv_ref spv_ref)¶ This subroutine computes the in situ specific volume of sea water (specvol in [m3 kg-1]) from salinity (S [PSU]), potential temperature (T [degC]) and pressure [Pa], using the UNESCO (1981) equation of state. If spv_ref is present, specvol is an anomaly from spv_ref.
- Parameters
[in] t: potential temperature relative to the surface [degC].[in] s: salinity [PSU].[in] pressure: pressure [Pa].[out] specvol: in situ specific volume [m3 kg-1].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.[in] spv_ref: A reference specific volume [m3 kg-1].
-
subroutine, public mom_eos_unesco::calculate_density_derivs_unesco(T T, S S, pressure pressure, drho_dT drho_dT, drho_dS drho_dS, start start, npts npts) This subroutine calculates the partial derivatives of density with potential temperature and salinity.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: Pressure [Pa].[out] drho_dt: The partial derivative of density with potential temperature [kg m-3 degC-1].[out] drho_ds: The partial derivative of density with salinity, in [kg m-3 PSU-1].[in] start: The starting point in the arrays.[in] npts: The number of values to calculate.
-
subroutine, public mom_eos_unesco::calculate_compress_unesco(T T, S S, pressure pressure, rho rho, drho_dp drho_dp, start start, npts npts) This subroutine computes the in situ density of sea water (rho) and the compressibility (drho/dp == C_sound^-2) at the given salinity, potential temperature, and pressure.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: Pressure [Pa].[out] rho: In situ density [kg m-3].[out] drho_dp: The partial derivative of density with pressure (also the inverse of the square of sound speed) [s2 m-2].[in] start: The starting point in the arrays.[in] npts: The number of values to calculate.
-
-
namespace
mom_eos_wright¶ The equation of state using the Wright 1997 expressions.
Unnamed Group
-
real, parameter mom_eos_wright::a0 = 7.057924e-4 Parameters in the Wright equation of state.
-
real, parameter mom_eos_wright::a1 = 3.480336e-7 Parameters in the Wright equation of state.
-
real, parameter mom_eos_wright::a2 = -1.112733e-7 Parameters in the Wright equation of state.
-
real, parameter mom_eos_wright::b0 = 5.790749e8 Parameters in the Wright equation of state.
-
real, parameter mom_eos_wright::b1 = 3.516535e6 Parameters in the Wright equation of state.
-
real, parameter mom_eos_wright::b2 = -4.002714e4 Parameters in the Wright equation of state.
-
real, parameter mom_eos_wright::b3 = 2.084372e2 Parameters in the Wright equation of state.
-
real, parameter mom_eos_wright::b4 = 5.944068e5 Parameters in the Wright equation of state.
-
real, parameter mom_eos_wright::b5 = -9.643486e3 Parameters in the Wright equation of state.
-
real, parameter mom_eos_wright::c0 = 1.704853e5 Parameters in the Wright equation of state.
-
real, parameter mom_eos_wright::c1 = 7.904722e2 Parameters in the Wright equation of state.
-
real, parameter mom_eos_wright::c2 = -7.984422 Parameters in the Wright equation of state.
-
real, parameter mom_eos_wright::c3 = 5.140652e-2 Parameters in the Wright equation of state.
-
real, parameter mom_eos_wright::c4 = -2.302158e2 Parameters in the Wright equation of state.
-
real, parameter mom_eos_wright::c5 = -3.079464 Parameters in the Wright equation of state.
-
subroutine
calculate_density_scalar_wright(T T, S S, pressure pressure, rho rho, rho_ref rho_ref)¶ This subroutine computes the in situ density of sea water (rho in [kg m-3]) from salinity (S [PSU]), potential temperature (T [degC]), and pressure [Pa]. It uses the expression from Wright, 1997, J. Atmos. Ocean. Tech., 14, 735-740.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: pressure [Pa].[out] rho: In situ density [kg m-3].[in] rho_ref: A reference density [kg m-3].
-
subroutine
calculate_density_array_wright(T T, S S, pressure pressure, rho rho, start start, npts npts, rho_ref rho_ref)¶ This subroutine computes the in situ density of sea water (rho in [kg m-3]) from salinity (S [PSU]), potential temperature (T [degC]), and pressure [Pa]. It uses the expression from Wright, 1997, J. Atmos. Ocean. Tech., 14, 735-740.
- Parameters
[in] t: potential temperature relative to the surface [degC].[in] s: salinity [PSU].[in] pressure: pressure [Pa].[out] rho: in situ density [kg m-3].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.[in] rho_ref: A reference density [kg m-3].
-
subroutine
calculate_spec_vol_scalar_wright(T T, S S, pressure pressure, specvol specvol, spv_ref spv_ref)¶ This subroutine computes the in situ specific volume of sea water (specvol in [m3 kg-1]) from salinity (S [PSU]), potential temperature (T [degC]) and pressure [Pa]. It uses the expression from Wright, 1997, J. Atmos. Ocean. Tech., 14, 735-740. If spv_ref is present, specvol is an anomaly from spv_ref.
- Parameters
[in] t: potential temperature relative to the surface [degC].[in] s: salinity [PSU].[in] pressure: pressure [Pa].[out] specvol: in situ specific volume [m3 kg-1].[in] spv_ref: A reference specific volume [m3 kg-1].
-
subroutine
calculate_spec_vol_array_wright(T T, S S, pressure pressure, specvol specvol, start start, npts npts, spv_ref spv_ref)¶ This subroutine computes the in situ specific volume of sea water (specvol in [m3 kg-1]) from salinity (S [PSU]), potential temperature (T [degC]) and pressure [Pa]. It uses the expression from Wright, 1997, J. Atmos. Ocean. Tech., 14, 735-740. If spv_ref is present, specvol is an anomaly from spv_ref.
- Parameters
[in] t: potential temperature relative to the surface [degC].[in] s: salinity [PSU].[in] pressure: pressure [Pa].[out] specvol: in situ specific volume [m3 kg-1].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.[in] spv_ref: A reference specific volume [m3 kg-1].
-
subroutine
calculate_density_derivs_array_wright(T T, S S, pressure pressure, drho_dT drho_dT, drho_dS drho_dS, start start, npts npts)¶ For a given thermodynamic state, return the thermal/haline expansion coefficients.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: pressure [Pa].[out] drho_dt: The partial derivative of density with potential temperature [kg m-3 degC-1].[out] drho_ds: The partial derivative of density with salinity, in [kg m-3 PSU-1].[in] start: The starting point in the arrays.[in] npts: The number of values to calculate.
-
subroutine
calculate_density_derivs_scalar_wright(T T, S S, pressure pressure, drho_dT drho_dT, drho_dS drho_dS)¶ The scalar version of calculate_density_derivs which promotes scalar inputs to a 1-element array and then demotes the output back to a scalar.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: pressure [Pa].[out] drho_dt: The partial derivative of density with potential temperature [kg m-3 degC-1].[out] drho_ds: The partial derivative of density with salinity, in [kg m-3 PSU-1].
-
subroutine
calculate_density_second_derivs_array_wright(T T, S S, P P, drho_ds_ds drho_ds_ds, drho_ds_dt drho_ds_dt, drho_dt_dt drho_dt_dt, drho_ds_dp drho_ds_dp, drho_dt_dp drho_dt_dp, start start, npts npts)¶ Second derivatives of density with respect to temperature, salinity, and pressure.
- Parameters
[in] t: Potential temperature referenced to 0 dbar [degC][in] s: Salinity [PSU][in] p: Pressure [Pa][out] drho_ds_ds: Partial derivative of beta with respect to S [kg m-3 PSU-2][out] drho_ds_dt: Partial derivative of beta with respcct to T [kg m-3 PSU-1 degC-1][out] drho_dt_dt: Partial derivative of alpha with respect to T [kg m-3 degC-2][out] drho_ds_dp: Partial derivative of beta with respect to pressure [kg m-3 PSU-1 Pa-1][out] drho_dt_dp: Partial derivative of alpha with respect to pressure [kg m-3 degC-1 Pa-1][in] start: Starting index in T,S,P[in] npts: Number of points to loop over
-
subroutine
calculate_density_second_derivs_scalar_wright(T T, S S, P P, drho_ds_ds drho_ds_ds, drho_ds_dt drho_ds_dt, drho_dt_dt drho_dt_dt, drho_ds_dp drho_ds_dp, drho_dt_dp drho_dt_dp)¶ Second derivatives of density with respect to temperature, salinity, and pressure for scalar inputs. Inputs promoted to 1-element array and output demoted to scalar.
- Parameters
[in] t: Potential temperature referenced to 0 dbar[in] s: Salinity [PSU][in] p: pressure [Pa][out] drho_ds_ds: Partial derivative of beta with respect to S [kg m-3 PSU-2][out] drho_ds_dt: Partial derivative of beta with respcct to T [kg m-3 PSU-1 degC-1][out] drho_dt_dt: Partial derivative of alpha with respect to T [kg m-3 degC-2][out] drho_ds_dp: Partial derivative of beta with respect to pressure [kg m-3 PSU-1 Pa-1][out] drho_dt_dp: Partial derivative of alpha with respect to pressure [kg m-3 degC-1 Pa-1]
-
subroutine, public mom_eos_wright::calculate_specvol_derivs_wright(T T, S S, pressure pressure, dSV_dT dSV_dT, dSV_dS dSV_dS, start start, npts npts) For a given thermodynamic state, return the partial derivatives of specific volume with temperature and salinity.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: pressure [Pa].[out] dsv_dt: The partial derivative of specific volume with potential temperature [m3 kg-1 degC-1].[out] dsv_ds: The partial derivative of specific volume with salinity [m3 kg-1 / Pa].[in] start: The starting point in the arrays.[in] npts: The number of values to calculate.
-
subroutine, public mom_eos_wright::calculate_compress_wright(T T, S S, pressure pressure, rho rho, drho_dp drho_dp, start start, npts npts) This subroutine computes the in situ density of sea water (rho in [kg m-3]) and the compressibility (drho/dp = C_sound^-2) (drho_dp [s2 m-2]) from salinity (sal in psu), potential temperature (T [degC]), and pressure [Pa]. It uses the expressions from Wright, 1997, J. Atmos. Ocean. Tech., 14, 735-740. Coded by R. Hallberg, 1/01.
- Parameters
[in] t: Potential temperature relative to the surface [degC].[in] s: Salinity [PSU].[in] pressure: pressure [Pa].[out] rho: In situ density [kg m-3].[out] drho_dp: The partial derivative of density with pressure (also the inverse of the square of sound speed) [s2 m-2].[in] start: The starting point in the arrays.[in] npts: The number of values to calculate.
-
subroutine, public mom_eos_wright::int_density_dz_wright(T T, S S, z_t z_t, z_b z_b, rho_ref rho_ref, rho_0 rho_0, G_e G_e, HII HII, HIO HIO, dpa dpa, intz_dpa intz_dpa, intx_dpa intx_dpa, inty_dpa inty_dpa, bathyT bathyT, dz_neglect dz_neglect, useMassWghtInterp useMassWghtInterp) This subroutine calculates analytical and nearly-analytical integrals of pressure anomalies across layers, which are required for calculating the finite-volume form pressure accelerations in a Boussinesq model.
- Parameters
[in] hii: The horizontal index type for the input arrays.[in] hio: The horizontal index type for the output arrays.[in] t: Potential temperature relative to the surface[in] s: Salinity [PSU].[in] z_t: Height at the top of the layer in depth units [Z ~> m].[in] z_b: Height at the top of the layer [Z ~> m].[in] rho_ref: A mean density [kg m-3], that is subtracted out to reduce the magnitude of each of the integrals. (The pressure is calucated as p~=-z*rho_0*G_e.)[in] rho_0: Density [kg m-3], that is used to calculate the pressure (as p~=-z*rho_0*G_e) used in the equation of state.[in] g_e: The Earth’s gravitational acceleration [m2 Z-1 s-2 ~> m s-2].[out] dpa: The change in the pressure anomaly across the[out] intz_dpa: The integral through the thickness of the layer[out] intx_dpa: The integral in x of the difference between the[out] inty_dpa: The integral in y of the difference between the[in] bathyt: The depth of the bathymetry [Z ~> m].[in] dz_neglect: A miniscule thickness change [Z ~> m].[in] usemasswghtinterp: If true, uses mass weighting to interpolate T/S for top and bottom integrals.
-
subroutine, public mom_eos_wright::int_spec_vol_dp_wright(T T, S S, p_t p_t, p_b p_b, spv_ref spv_ref, HI HI, dza dza, intp_dza intp_dza, intx_dza intx_dza, inty_dza inty_dza, halo_size halo_size, bathyP bathyP, dP_neglect dP_neglect, useMassWghtInterp useMassWghtInterp) This subroutine calculates analytical and nearly-analytical integrals in pressure across layers of geopotential anomalies, which are required for calculating the finite-volume form pressure accelerations in a non-Boussinesq model. There are essentially no free assumptions, apart from the use of Bode’s rule to do the horizontal integrals, and from a truncation in the series for log(1-eps/1+eps) that assumes that |eps| < 0.34.
- Parameters
[in] hi: The ocean’s horizontal index type.[in] t: Potential temperature relative to the surface[in] s: Salinity [PSU].[in] p_t: Pressure at the top of the layer [Pa].[in] p_b: Pressure at the top of the layer [Pa].[in] spv_ref: A mean specific volume that is subtracted out to reduce the magnitude of each of the integrals [m3 kg-1]. The calculation is mathematically identical with different values of spv_ref, but this reduces the effects of roundoff.[out] dza: The change in the geopotential anomaly across[out] intp_dza: The integral in pressure through the layer of[out] intx_dza: The integral in x of the difference between the[out] inty_dza: The integral in y of the difference between the[in] halo_size: The width of halo points on which to calculate dza.[in] bathyp: The pressure at the bathymetry [Pa][in] dp_neglect: A miniscule pressure change with the same units as p_t [Pa][in] usemasswghtinterp: If true, uses mass weighting to interpolate T/S for top and bottom integrals.
-
-
namespace
mom_error_handler¶ Routines for error handling and I/O management.
Functions
-
logical function, public mom_error_handler::is_root_pe() This returns .true. if the current PE is the root PE.
-
subroutine, public mom_error_handler::mom_mesg(message message, verb verb, all_print all_print) This provides a convenient interface for writing an informative comment.
- Parameters
[in] message: A message to write out[in] verb: A level of verbosity for this message[in] all_print: If present and true, any PEs are able to write this message.
-
subroutine, public mom_error_handler::mom_error(level level, message message, all_print all_print) This provides a convenient interface for writing an mpp_error message with run-time filter based on a verbosity.
- Parameters
[in] level: The verbosity level of this message[in] message: A message to write out[in] all_print: If present and true, any PEs are able to write this message.
-
subroutine, public mom_error_handler::mom_set_verbosity(verb verb) This subroutine sets the level of verbosity filtering MOM error messages.
- Parameters
[in] verb: A level of verbosity to set
-
integer function, public mom_error_handler::mom_get_verbosity() This subroutine gets the level of verbosity filtering MOM error messages.
-
logical function, public mom_error_handler::mom_verbose_enough(verb verb) This tests whether the level of verbosity filtering MOM error messages is sufficient to write a message of verbosity level verb.
- Parameters
[in] verb: A level of verbosity to test
-
logical function, public mom_error_handler::calltree_showquery() Returns True, if the verbosity>=6 indicating to show the call tree.
-
subroutine, public mom_error_handler::calltree_enter(mesg mesg, n n) Writes a message about entering a subroutine if call tree reporting is active.
- Parameters
[in] mesg: Message to write[in] n: An optional integer to write at end of message
-
subroutine, public mom_error_handler::calltree_leave(mesg mesg) Writes a message about leaving a subroutine if call tree reporting is active.
- Parameters
mesg: Message to write
-
subroutine, public mom_error_handler::calltree_waypoint(mesg mesg, n n) Writes a message about reaching a milestone if call tree reporting is active.
- Parameters
[in] mesg: Message to write[in] n: An optional integer to write at end of message
-
subroutine, public mom_error_handler::assert(logical_arg logical_arg, msg msg) Issues a FATAL error if the assertion fails, i.e. the first argument is false.
- Parameters
[in] logical_arg: If false causes a FATAL error[in] msg: Message to issue in case of failed assertion
Variables
-
integer
verbosity= 6¶ Verbosity level: 0 - FATAL messages only 1 - FATAL + WARNING messages only 2 - FATAL + WARNING + NOTE messages only [default] 3 - above + informational 4 - 5 - 6 - above + call tree 7 - 8 - 9 - anything and everything (also set with DEBUG=True)
-
integer
calltreeindentlevel= 0¶ The level of calling within the call tree.
-
-
namespace
mom_file_parser¶ The MOM6 facility to parse input files for runtime parameters.
By Robert Hallberg and Alistair Adcroft, updated 9/2013.
The subroutines here parse a set of input files for the value a named parameter and sets that parameter at run time. Currently these files use use one of several formats: #define VAR ! To set the logical VAR to true. VAR = True ! To set the logical VAR to true. #undef VAR ! To set the logical VAR to false. VAR = False ! To set the logical VAR to false. #define VAR 999 ! To set the real or integer VAR to 999. VAR = 999 ! To set the real or integer VAR to 999. #override VAR = 888 ! To override a previously set value. VAR = 1.1, 2.2, 3.3 ! To set an array of real values.
Unnamed Group
-
logical, parameter mom_file_parser::report_unused_default = .false. Default values for parameters.
-
logical, parameter mom_file_parser::unused_params_fatal_default = .false. Default values for parameters.
-
logical, parameter mom_file_parser::log_to_stdout_default = .false. Default values for parameters.
-
logical, parameter mom_file_parser::complete_doc_default = .true. Default values for parameters.
-
logical, parameter mom_file_parser::minimal_doc_default = .true. Default values for parameters.
-
subroutine, public mom_file_parser::open_param_file(filename filename, CS CS, checkable checkable, component component, doc_file_dir doc_file_dir) Make the contents of a parameter input file availalble in a param_file_type.
- Parameters
[in] filename: An input file name, optionally with the full path[inout] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] checkable: If this is false, it disables checks of this file for unused parameters. The default is True.[in] component: If present, this component name is used to generate parameter documentation file names; the default is”MOM”[in] doc_file_dir: An optional directory in which to write out the documentation files. The default is effectively ‘./’.
-
subroutine, public mom_file_parser::close_param_file(CS CS, quiet_close quiet_close, component component) Close any open input files and deallocate memory associated with this param_file_type. To use this type again, open_param_file would have to be called again.
- Parameters
[inout] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] quiet_close: if present and true, do not do any logging with this call.[in] component: If present, this component name is used to generate parameter documentation file names
-
subroutine
populate_param_data(iounit iounit, filename filename, param_data param_data)¶ Read the contents of a parameter input file, and store the contents in a file_data_type after removing comments and simplifying white space.
- Parameters
[in] iounit: The IO unit number that is open for filename[in] filename: An input file name, optionally with the full path[inout] param_data: A list of the input lines that set parameters after comments have been stripped out.
-
logical function mom_file_parser::openmultilinecomment(string string) Return True if a /* appears on this line without a closing */.
- Parameters
[in] string: The input string to process
-
logical function mom_file_parser::closemultilinecomment(string string) Return True if a */ appears on this line.
- Parameters
[in] string: The input string to process
-
integer function mom_file_parser::lastnoncommentindex(string string) Find position of last character before any comments, As marked by “!”, “//”, or “/*” following F90, C++, or C syntax.
- Parameters
[in] string: The input string to process
-
integer function mom_file_parser::lastnoncommentnonblank(string string) Find position of last non-blank character before any comments.
- Parameters
[in] string: The input string to process
-
character(len=len(string)) function mom_file_parser::replacetabs(string string) Returns a string with tabs replaced by a blank.
- Parameters
[in] string: The input string to process
-
character(len=len(string)) function mom_file_parser::removecomments(string string) Trims comments and leading blanks from string.
- Parameters
[in] string: The input string to process
-
character(len=len(string)+16) function mom_file_parser::simplifywhitespace(string string) Constructs a string with all repeated whitespace replaced with single blanks and insert white space where it helps delineate tokens (e.g. around =)
- Parameters
[in] string: A string to modify to simpify white space
-
subroutine
read_param_int(CS CS, varname varname, value value, fail_if_missing fail_if_missing)¶ This subroutine reads the value of an integer model parameter from a parameter file.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file
-
subroutine
read_param_int_array(CS CS, varname varname, value value, fail_if_missing fail_if_missing)¶ This subroutine reads the values of an array of integer model parameters from a parameter file.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file
-
subroutine
read_param_real(CS CS, varname varname, value value, fail_if_missing fail_if_missing, scale scale)¶ This subroutine reads the value of a real model parameter from a parameter file.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file[in] scale: A scaling factor that the parameter is multiplied by before it is returned.
-
subroutine
read_param_real_array(CS CS, varname varname, value value, fail_if_missing fail_if_missing, scale scale)¶ This subroutine reads the values of an array of real model parameters from a parameter file.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file[in] scale: A scaling factor that the parameter is multiplied by before it is returned.
-
subroutine
read_param_char(CS CS, varname varname, value value, fail_if_missing fail_if_missing)¶ This subroutine reads the value of a character string model parameter from a parameter file.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file
-
subroutine
read_param_char_array(CS CS, varname varname, value value, fail_if_missing fail_if_missing)¶ This subroutine reads the values of an array of character string model parameters from a parameter file.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file
-
subroutine
read_param_logical(CS CS, varname varname, value value, fail_if_missing fail_if_missing)¶ This subroutine reads the value of a logical model parameter from a parameter file.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file
-
subroutine
read_param_time(CS CS, varname varname, value value, timeunit timeunit, fail_if_missing fail_if_missing, date_format date_format)¶ This subroutine reads the value of a time_type model parameter from a parameter file.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file[in] timeunit: The number of seconds in a time unit for real-number input.[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file[out] date_format: If present, this indicates whether this parameter was read in a date format, so that it can later be logged in the same format.
-
character(len=input_str_length) function mom_file_parser::strip_quotes(val_str val_str) This function removes single and double quotes from a character string.
- Parameters
val_str: The character string to work on
-
subroutine
get_variable_line(CS CS, varname varname, found found, defined defined, value_string value_string, paramIsLogical paramIsLogical)¶ This subtoutine extracts the contents of lines in the param_file_type that refer to a named parameter. The value_string that is returned must be interepreted in a way that depends on the type of this variable.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] varname: The case-sensitive name of the parameter to read[out] found: If true, this parameter has been found in CS[out] defined: If true, this parameter is set (or true) in the CS[out] value_string: A string that encodes the new value[in] paramislogical: If true, this is a logical parameter that can be simply defined without parsing a value_string.
-
subroutine
flag_line_as_read(line_used line_used, count count)¶ Record that a line has been used to set a parameter.
- Parameters
line_used: A structure indicating which lines have been read[in] count: The parameter on this line number has been read
-
logical function mom_file_parser::overridewarninghasbeenissued(chain chain, varName varName) Returns true if an override warning has been issued for the variable varName.
- Parameters
chain: The linked list of variables that have already had override warnings issued[in] varname: The name of the variable being queried for warnings
-
subroutine
log_version_cs(CS CS, modulename modulename, version version, desc desc)¶ Log the version of a module to a log file and/or stdout, and/or to the parameter documentation file.
- Parameters
[in] cs: File parser type[in] modulename: Name of calling module[in] version: Version string of module[in] desc: Module description
-
subroutine
log_version_plain(modulename modulename, version version)¶ Log the version of a module to a log file and/or stdout.
- Parameters
[in] modulename: Name of calling module[in] version: Version string of module
-
subroutine mom_file_parser::log_param_int(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, layoutParam layoutParam, debuggingParam debuggingParam) Log the name and value of an integer model parameter in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the module using this parameter[in] varname: The name of the parameter to log[in] value: The value of the parameter to log[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] layoutparam: If present and true, this parameter is logged in the layout parameter file[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file
-
subroutine mom_file_parser::log_param_int_array(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, layoutParam layoutParam, debuggingParam debuggingParam) Log the name and values of an array of integer model parameter in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the module using this parameter[in] varname: The name of the parameter to log[in] value: The value of the parameter to log[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] layoutparam: If present and true, this parameter is logged in the layout parameter file[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file
-
subroutine mom_file_parser::log_param_real(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, debuggingParam debuggingParam) Log the name and value of a real model parameter in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The name of the parameter to log[in] value: The value of the parameter to log[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file
-
subroutine mom_file_parser::log_param_real_array(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, debuggingParam debuggingParam) Log the name and values of an array of real model parameter in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The name of the parameter to log[in] value: The value of the parameter to log[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file
-
subroutine mom_file_parser::log_param_logical(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, layoutParam layoutParam, debuggingParam debuggingParam) Log the name and value of a logical model parameter in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The name of the parameter to log[in] value: The value of the parameter to log[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] layoutparam: If present and true, this parameter is logged in the layout parameter file[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file
-
subroutine mom_file_parser::log_param_char(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, layoutParam layoutParam, debuggingParam debuggingParam) Log the name and value of a character string model parameter in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The name of the parameter to log[in] value: The value of the parameter to log[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] layoutparam: If present and true, this parameter is logged in the layout parameter file[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file
-
subroutine mom_file_parser::log_param_time(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, timeunit timeunit, layoutParam layoutParam, debuggingParam debuggingParam, log_date log_date) This subroutine writes the value of a time-type parameter to a log file, along with its name and the module it came from.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The name of the parameter to log[in] value: The value of the parameter to log[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] timeunit: The number of seconds in a time unit for real-number output.[in] log_date: If true, log the time_type in date format. If missing the default is false.[in] layoutparam: If present and true, this parameter is logged in the layout parameter file[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file
-
character(len=40) function mom_file_parser::convert_date_to_string(date date) This function converts a date into a string, valid with ticks and for dates up to year 99,999,999.
- Return
A date string in a format like YYYY-MM-DD HH:MM:SS.sss
- Parameters
[in] date: The date to be translated into a string.
-
subroutine mom_file_parser::get_param_int(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, fail_if_missing fail_if_missing, do_not_read do_not_read, do_not_log do_not_log, static_value static_value, layoutParam layoutParam, debuggingParam debuggingParam) This subroutine reads the value of an integer model parameter from a parameter file and logs it in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file and logged[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] static_value: If this parameter is static, it takes this value, which can be compared for consistency with what is in the parameter file.[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file[in] do_not_read: If present and true, do not read a value for this parameter, although it might be logged.[in] do_not_log: If present and true, do not log this parameter to the documentation files[in] layoutparam: If present and true, this parameter is logged in the layout parameter file[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file
-
subroutine mom_file_parser::get_param_int_array(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, fail_if_missing fail_if_missing, do_not_read do_not_read, do_not_log do_not_log, static_value static_value, layoutParam layoutParam, debuggingParam debuggingParam) This subroutine reads the values of an array of integer model parameters from a parameter file and logs them in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be reset from the parameter file[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] static_value: If this parameter is static, it takes this value, which can be compared for consistency with what is in the parameter file.[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file[in] do_not_read: If present and true, do not read a value for this parameter, although it might be logged.[in] do_not_log: If present and true, do not log this parameter to the documentation files[in] layoutparam: If present and true, this parameter is logged in the layout parameter file[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file
-
subroutine mom_file_parser::get_param_real(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, fail_if_missing fail_if_missing, do_not_read do_not_read, do_not_log do_not_log, static_value static_value, debuggingParam debuggingParam, scale scale, unscaled unscaled) This subroutine reads the value of a real model parameter from a parameter file and logs it in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file and logged[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] static_value: If this parameter is static, it takes this value, which can be compared for consistency with what is in the parameter file.[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file[in] do_not_read: If present and true, do not read a value for this parameter, although it might be logged.[in] do_not_log: If present and true, do not log this parameter to the documentation files[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file[in] scale: A scaling factor that the parameter is multiplied by before it is returned.[out] unscaled: The value of the parameter that would be returned without any multiplication by a scaling factor.
-
subroutine mom_file_parser::get_param_real_array(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, fail_if_missing fail_if_missing, do_not_read do_not_read, do_not_log do_not_log, debuggingParam debuggingParam, static_value static_value, scale scale, unscaled unscaled) This subroutine reads the values of an array of real model parameters from a parameter file and logs them in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file and logged[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] static_value: If this parameter is static, it takes this value, which can be compared for consistency with what is in the parameter file.[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file[in] do_not_read: If present and true, do not read a value for this parameter, although it might be logged.[in] do_not_log: If present and true, do not log this parameter to the documentation files[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file[in] scale: A scaling factor that the parameter is multiplied by before it is returned.[out] unscaled: The value of the parameter that would be returned without any multiplication by a scaling factor.
-
subroutine mom_file_parser::get_param_char(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, fail_if_missing fail_if_missing, do_not_read do_not_read, do_not_log do_not_log, static_value static_value, layoutParam layoutParam, debuggingParam debuggingParam) This subroutine reads the value of a character string model parameter from a parameter file and logs it in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file and logged[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] static_value: If this parameter is static, it takes this value, which can be compared for consistency with what is in the parameter file.[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file[in] do_not_read: If present and true, do not read a value for this parameter, although it might be logged.[in] do_not_log: If present and true, do not log this parameter to the documentation files[in] layoutparam: If present and true, this parameter is logged in the layout parameter file[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file
-
subroutine mom_file_parser::get_param_char_array(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, fail_if_missing fail_if_missing, do_not_read do_not_read, do_not_log do_not_log, static_value static_value) This subroutine reads the values of an array of character string model parameters from a parameter file and logs them in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file and logged[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] static_value: If this parameter is static, it takes this value, which can be compared for consistency with what is in the parameter file.[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file[in] do_not_read: If present and true, do not read a value for this parameter, although it might be logged.[in] do_not_log: If present and true, do not log this parameter to the documentation files
-
subroutine mom_file_parser::get_param_logical(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, fail_if_missing fail_if_missing, do_not_read do_not_read, do_not_log do_not_log, static_value static_value, layoutParam layoutParam, debuggingParam debuggingParam) This subroutine reads the value of a logical model parameter from a parameter file and logs it in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file and logged[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] static_value: If this parameter is static, it takes this value, which can be compared for consistency with what is in the parameter file.[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file[in] do_not_read: If present and true, do not read a value for this parameter, although it might be logged.[in] do_not_log: If present and true, do not log this parameter to the documentation files[in] layoutparam: If present and true, this parameter is logged in the layout parameter file[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file
-
subroutine mom_file_parser::get_param_time(CS CS, modulename modulename, varname varname, value value, desc desc, units units, default default, fail_if_missing fail_if_missing, do_not_read do_not_read, do_not_log do_not_log, timeunit timeunit, static_value static_value, layoutParam layoutParam, debuggingParam debuggingParam, log_as_date log_as_date) This subroutine reads the value of a time-type model parameter from a parameter file and logs it in documentation files.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] modulename: The name of the calling module[in] varname: The case-sensitive name of the parameter to read[inout] value: The value of the parameter that may be read from the parameter file and logged[in] desc: A description of this variable; if not present, this parameter is not written to a doc file[in] units: The units of this parameter[in] default: The default value of the parameter[in] static_value: If this parameter is static, it takes this value, which can be compared for consistency with what is in the parameter file.[in] fail_if_missing: If present and true, a fatal error occurs if this variable is not found in the parameter file[in] do_not_read: If present and true, do not read a value for this parameter, although it might be logged.[in] do_not_log: If present and true, do not log this parameter to the documentation files[in] timeunit: The number of seconds in a time unit for real-number input to be translated to a time.[in] layoutparam: If present and true, this parameter is logged in the layout parameter file[in] debuggingparam: If present and true, this parameter is logged in the debugging parameter file[in] log_as_date: If true, log the time_type in date format. The default is false.
-
subroutine, public mom_file_parser::clearparameterblock(CS CS) Resets the parameter block name to blank.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters
-
subroutine, public mom_file_parser::openparameterblock(CS CS, blockName blockName, desc desc) Tags blockName onto the end of the active parameter block name.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters[in] blockname: The name of a parameter block being added[in] desc: A description of the parameter block being added
-
subroutine, public mom_file_parser::closeparameterblock(CS CS) Remove the lowest level of recursion from the active block name.
- Parameters
[in] cs: The control structure for the file_parser module, it is also a structure to parse for run-time parameters
-
character(len=len(oldblockname)+40) function mom_file_parser::pushblocklevel(oldblockName oldblockName, newBlockName newBlockName) Extends block name (deeper level of parameter block)
- Parameters
[in] oldblockname: A sequence of hierarchical parameter block names[in] newblockname: A new block name to add to the end of the sequence
-
character(len=len(oldblockname)+40) function mom_file_parser::popblocklevel(oldblockName oldblockName) Truncates block name (shallower level of parameter block)
- Parameters
[in] oldblockname: A sequence of hierarchical parameter block names
Variables
-
integer, parameter, public mom_file_parser::max_param_files = 5 Maximum number of parameter files.
-
integer, parameter mom_file_parser::input_str_length = 320 Maximum line length in parameter file.
-
integer, parameter mom_file_parser::filename_length = 200 Maximum number of characters in file names.
-
logical
all_pes_read= .false.¶ If true, all PEs read the input files TODO: Eliminate this parameter.
-
-
namespace
mom_fixed_initialization¶ Initializes fixed aspects of the model, such as horizontal grid metrics, topography and Coriolis.
Functions
-
subroutine, public mom_fixed_initialization::mom_initialize_fixed(G G, US US, OBC OBC, PF PF, write_geom write_geom, output_dir output_dir) MOM_initialize_fixed sets up time-invariant quantities related to MOM6’s horizontal grid, bathymetry, and the Coriolis parameter.
- Parameters
[inout] g: The ocean’s grid structure.[in] us: A dimensional unit scaling typeobc: Open boundary structure.[in] pf: A structure indicating the open file to parse for model parameter values.[in] write_geom: If true, write grid geometry files.[in] output_dir: The directory into which to write files.
-
subroutine, public mom_fixed_initialization::mom_initialize_topography(D D, max_depth max_depth, G G, PF PF, US US) MOM_initialize_topography makes the appropriate call to set up the bathymetry. At this point the topography is in units of [m], but this can be changed later.
- Parameters
[in] g: The dynamic horizontal grid type[out] d: Ocean bottom depth [m][in] pf: Parameter file structure[out] max_depth: Maximum depth of model [m][in] us: A dimensional unit scaling type
-
-
namespace
mom_forcing_type¶ This module implements boundary forcing for MOM6.
Unnamed Group
-
subroutine, public mom_forcing_type::extractfluxes1d(G G, GV GV, fluxes fluxes, optics optics, nsw nsw, j j, dt dt, FluxRescaleDepth FluxRescaleDepth, useRiverHeatContent useRiverHeatContent, useCalvingHeatContent useCalvingHeatContent, h h, T T, netMassInOut netMassInOut, netMassOut netMassOut, net_heat net_heat, net_salt net_salt, pen_SW_bnd pen_SW_bnd, tv tv, aggregate_FW aggregate_FW, nonpenSW nonpenSW, netmassInOut_rate netmassInOut_rate, net_Heat_Rate net_Heat_Rate, net_salt_rate net_salt_rate, pen_sw_bnd_Rate pen_sw_bnd_Rate, skip_diags skip_diags) This subroutine extracts fluxes from the surface fluxes type. It works on a j-row for optimization purposes. The 2d (i,j) wrapper is the next subroutine below. This routine multiplies fluxes by dt, so that the result is an accumulation of fluxes over a time step.
- Parameters
[in] g: ocean grid structure[in] gv: ocean vertical grid structure[inout] fluxes: structure containing pointers to possible forcing fields. NULL unused fields.optics: pointer to optics[in] nsw: number of bands of penetrating SW[in] j: j-index to work on[in] dt: time step [s][in] fluxrescaledepth: min ocean depth before fluxes are scaled away [H ~> m or kg m-2][in] useriverheatcontent: logical for river heat content[in] usecalvingheatcontent: logical for calving heat content[in] h: layer thickness [H ~> m or kg m-2][in] t: layer temperatures [degC][out] netmassinout: net mass flux (non-Bouss) or volume flux (if Bouss) of water in/out of ocean over a time step [H ~> m or kg m-2][out] netmassout: net mass flux (non-Bouss) or volume flux (if Bouss) of water leaving ocean surface over a time step [H ~> m or kg m-2]. netMassOut < 0 means mass leaves ocean.[out] net_heat: net heat at the surface accumulated over a time step for coupler + restoring. Exclude two terms from net_heat: (1) downwelling (penetrative) SW, (2) evaporation heat content, (since do not yet know evap temperature). [degC H ~> degC m or degC kg m-2].[out] net_salt: surface salt flux into the ocean accumulated over a time step [ppt H ~> ppt m or ppt kg m-2].[out] pen_sw_bnd: penetrating SW flux, split into bands. [degC H ~> degC m or degC kg m-2] and array size nsw x G isd: G ied, where nsw=number of SW bands in pen_SW_bnd. This heat flux is not part of net_heat.[inout] tv: structure containing pointers to available thermodynamic fields. Used to keep track of the heat flux associated with net mass fluxes into the ocean.[in] aggregate_fw: For determining how to aggregate forcing.[out] nonpensw: Non-penetrating SW used in net_heat[out] net_heat_rate: Rate of net surface heating[out] net_salt_rate: Surface salt flux into the ocean[out] netmassinout_rate: Rate of net mass flux into the ocean[out] pen_sw_bnd_rate: Rate of penetrative shortwave heating[in] skip_diags: If present and true, skip calculating diagnostics
-
subroutine, public mom_forcing_type::extractfluxes2d(G G, GV GV, fluxes fluxes, optics optics, nsw nsw, dt dt, FluxRescaleDepth FluxRescaleDepth, useRiverHeatContent useRiverHeatContent, useCalvingHeatContent useCalvingHeatContent, h h, T T, netMassInOut netMassInOut, netMassOut netMassOut, net_heat net_heat, Net_salt Net_salt, Pen_SW_bnd Pen_SW_bnd, tv tv, aggregate_FW aggregate_FW) 2d wrapper for 1d extract fluxes from surface fluxes type. This subroutine extracts fluxes from the surface fluxes type. It multiplies the fluxes by dt, so that the result is an accumulation of the fluxes over a time step.
- Parameters
[in] g: ocean grid structure[in] gv: ocean vertical grid structure[inout] fluxes: structure containing pointers to forcing.optics: pointer to optics[in] nsw: number of bands of penetrating SW[in] dt: time step [s][in] fluxrescaledepth: min ocean depth before fluxes are scaled away [H ~> m or kg m-2][in] useriverheatcontent: logical for river heat content[in] usecalvingheatcontent: logical for calving heat content[in] h: layer thickness [H ~> m or kg m-2][in] t: layer temperatures [degC][out] netmassinout: net mass flux (non-Bouss) or volume flux (if Bouss) of water in/out of ocean over a time step [H ~> m or kg m-2][out] netmassout: net mass flux (non-Bouss) or volume flux (if Bouss) of water leaving ocean surface over a time step [H ~> m or kg m-2].[out] net_heat: net heat at the surface accumulated over a time step associated with coupler + restore. Exclude two terms from net_heat: (1) downwelling (penetrative) SW, (2) evaporation heat content, (since do not yet know temperature of evap). [degC H ~> degC m or degC kg m-2][out] net_salt: surface salt flux into the ocean accumulated over a time step [ppt H ~> ppt m or ppt kg m-2][out] pen_sw_bnd: penetrating SW flux, by frequency band [degC H ~> degC m or degC kg m-2] with array size nsw x G isd: G ied, where nsw=number of SW bands in pen_SW_bnd. This heat flux is not in net_heat.[inout] tv: structure containing pointers to available thermodynamic fields. Here it is used to keep track of the heat flux associated with net mass fluxes into the ocean.[in] aggregate_fw: For determining how to aggregate the forcing.
-
subroutine, public mom_forcing_type::calculatebuoyancyflux1d(G G, GV GV, US US, fluxes fluxes, optics optics, nsw nsw, h h, Temp Temp, Salt Salt, tv tv, j j, buoyancyFlux buoyancyFlux, netHeatMinusSW netHeatMinusSW, netSalt netSalt, skip_diags skip_diags) This routine calculates surface buoyancy flux by adding up the heat, FW & salt fluxes. These are actual fluxes, with units of stuff per time. Setting dt=1 in the call to extractFluxes routine allows us to get “stuf per time” rather than the time integrated fluxes needed in other routines that call extractFluxes.
- Parameters
[in] g: ocean grid[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] fluxes: surface fluxesoptics: penetrating SW optics[in] nsw: The number of frequency bands of penetrating shortwave radiation[in] h: layer thickness [H ~> m or kg m-2][in] temp: prognostic temp [degC][in] salt: salinity [ppt][inout] tv: thermodynamics type[in] j: j-row to work on[inout] buoyancyflux: buoyancy fluxes [L2 T-3 ~> m2 s-3][inout] netheatminussw: surf Heat flux [degC H s-1 ~> degC m s-1 or degC kg m-2 s-1][inout] netsalt: surf salt flux [ppt H s-1 ~> ppt m s-1 or ppt kg m-2 s-1][in] skip_diags: If present and true, skip calculating diagnostics inside extractFluxes1d()
-
subroutine, public mom_forcing_type::calculatebuoyancyflux2d(G G, GV GV, US US, fluxes fluxes, optics optics, h h, Temp Temp, Salt Salt, tv tv, buoyancyFlux buoyancyFlux, netHeatMinusSW netHeatMinusSW, netSalt netSalt, skip_diags skip_diags) Calculates surface buoyancy flux by adding up the heat, FW and salt fluxes, for 2d arrays. This is a wrapper for calculateBuoyancyFlux1d.
- Parameters
[in] g: ocean grid[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] fluxes: surface fluxesoptics: SW ocean optics[in] h: layer thickness [H ~> m or kg m-2][in] temp: temperature [degC][in] salt: salinity [ppt][inout] tv: thermodynamics type[inout] buoyancyflux: buoyancy fluxes [L2 T-3 ~> m2 s-3][inout] netheatminussw: surf temp flux [degC H ~> degC m or degC kg m-2][inout] netsalt: surf salt flux [ppt H ~> ppt m or ppt kg m-2][in] skip_diags: If present and true, skip calculating diagnostics inside extractFluxes1d()
-
subroutine, public mom_forcing_type::mom_forcing_chksum(mesg mesg, fluxes fluxes, G G, US US, haloshift haloshift) Write out chksums for thermodynamic fluxes.
- Parameters
[in] mesg: message[in] fluxes: A structure containing thermodynamic forcing fields[in] g: grid type[in] us: A dimensional unit scaling type[in] haloshift: shift in halo
-
subroutine, public mom_forcing_type::mom_mech_forcing_chksum(mesg mesg, forces forces, G G, US US, haloshift haloshift) Write out chksums for the driving mechanical forces.
- Parameters
[in] mesg: message[in] forces: A structure with the driving mechanical forces[in] g: grid type[in] us: A dimensional unit scaling type[in] haloshift: shift in halo
-
subroutine
mech_forcing_singlepointprint(forces forces, G G, i i, j j, mesg mesg)¶ Write out values of the mechanical forcing arrays at the i,j location. This is a debugging tool.
- Parameters
[in] forces: A structure with the driving mechanical forces[in] g: Grid type[in] mesg: Message[in] i: i-index[in] j: j-index
-
subroutine, public mom_forcing_type::forcing_singlepointprint(fluxes fluxes, G G, i i, j j, mesg mesg) Write out values of the fluxes arrays at the i,j location. This is a debugging tool.
- Parameters
[in] fluxes: A structure containing thermodynamic forcing fields[in] g: Grid type[in] mesg: Message[in] i: i-index[in] j: j-index
-
subroutine, public mom_forcing_type::register_forcing_type_diags(Time Time, diag diag, US US, use_temperature use_temperature, handles handles, use_berg_fluxes use_berg_fluxes) Register members of the forcing type for diagnostics.
- Parameters
[in] time: time type[inout] diag: diagnostic control type[in] us: A dimensional unit scaling type[in] use_temperature: True if T/S are in use[inout] handles: handles for diagnostics[in] use_berg_fluxes: If true, allow iceberg flux diagnostics
-
subroutine, public mom_forcing_type::forcing_accumulate(flux_tmp flux_tmp, forces forces, fluxes fluxes, dt dt, G G, wt2 wt2) Accumulate the forcing over time steps, taking input from a mechanical forcing type and a temporary forcing-flux type.
- Parameters
[in] flux_tmp: A temporary structure with current thermodynamic forcing fields[in] forces: A structure with the driving mechanical forces[inout] fluxes: A structure containing time-averaged thermodynamic forcing fields[in] dt: The elapsed time since the last call to this subroutine [s][inout] g: The ocean’s grid structure[out] wt2: The relative weight of the new fluxes
-
subroutine, public mom_forcing_type::fluxes_accumulate(flux_tmp flux_tmp, fluxes fluxes, dt dt, G G, wt2 wt2, forces forces) Accumulate the thermodynamic fluxes over time steps.
- Parameters
[in] flux_tmp: A temporary structure with current thermodynamic forcing fields[inout] fluxes: A structure containing time-averaged thermodynamic forcing fields[in] dt: The elapsed time since the last call to this subroutine [s][inout] g: The ocean’s grid structure[out] wt2: The relative weight of the new fluxes[in] forces: A structure with the driving mechanical forces
-
subroutine, public mom_forcing_type::copy_common_forcing_fields(forces forces, fluxes fluxes, G G, skip_pres skip_pres) This subroutine copies the computational domains of common forcing fields from a mech_forcing type to a (thermodynamic) forcing type.
- Parameters
[in] forces: A structure with the driving mechanical forces[inout] fluxes: A structure containing thermodynamic forcing fields[in] g: grid type[in] skip_pres: If present and true, do not copy pressure fields.
-
subroutine, public mom_forcing_type::set_derived_forcing_fields(forces forces, fluxes fluxes, G G, US US, Rho0 Rho0) This subroutine calculates certain derived forcing fields based on information from a mech_forcing type and stores them in a (thermodynamic) forcing type.
- Parameters
[in] forces: A structure with the driving mechanical forces[inout] fluxes: A structure containing thermodynamic forcing fields[in] g: grid type[in] us: A dimensional unit scaling type[in] rho0: A reference density of seawater [kg m-3], as used to calculate ustar.
-
subroutine, public mom_forcing_type::set_net_mass_forcing(fluxes fluxes, forces forces, G G) This subroutine determines the net mass source to the ocean from a (thermodynamic) forcing type and stores it in a mech_forcing type.
- Parameters
[in] fluxes: A structure containing thermodynamic forcing fields[inout] forces: A structure with the driving mechanical forces[in] g: The ocean grid type
-
subroutine, public mom_forcing_type::get_net_mass_forcing(fluxes fluxes, G G, net_mass_src net_mass_src) This subroutine calculates determines the net mass source to the ocean from a (thermodynamic) forcing type and stores it in a provided array.
- Parameters
[in] fluxes: A structure containing thermodynamic forcing fields[in] g: The ocean grid type[out] net_mass_src: The net mass flux of water into the ocean [kg m-2 s-1].
-
subroutine, public mom_forcing_type::copy_back_forcing_fields(fluxes fluxes, forces forces, G G) This subroutine copies the computational domains of common forcing fields from a mech_forcing type to a (thermodynamic) forcing type.
- Parameters
[in] fluxes: A structure containing thermodynamic forcing fields[inout] forces: A structure with the driving mechanical forces[in] g: grid type
-
subroutine, public mom_forcing_type::mech_forcing_diags(forces forces, dt dt, G G, diag diag, handles handles) Offer mechanical forcing fields for diagnostics for those fields registered as part of register_forcing_type_diags.
- Parameters
[in] forces: A structure with the driving mechanical forces[in] dt: time step[in] g: grid type[in] diag: diagnostic type[inout] handles: diagnostic id for diag_manager
-
subroutine, public mom_forcing_type::forcing_diagnostics(fluxes fluxes, sfc_state sfc_state, dt dt, G G, diag diag, handles handles) Offer buoyancy forcing fields for diagnostics for those fields registered as part of register_forcing_type_diags.
- Parameters
[in] fluxes: A structure containing thermodynamic forcing fields[in] sfc_state: A structure containing fields that describe the surface state of the ocean.[in] dt: time step[in] g: grid type[in] diag: diagnostic regulator[inout] handles: diagnostic ids
-
subroutine, public mom_forcing_type::allocate_forcing_type(G G, fluxes fluxes, water water, heat heat, ustar ustar, press press, shelf shelf, iceberg iceberg, salt salt) Conditionally allocate fields within the forcing type.
- Parameters
[in] g: Ocean grid structure[inout] fluxes: A structure containing thermodynamic forcing fields[in] water: If present and true, allocate water fluxes[in] heat: If present and true, allocate heat fluxes[in] ustar: If present and true, allocate ustar and related fields[in] press: If present and true, allocate p_surf and related fields[in] shelf: If present and true, allocate fluxes for ice-shelf[in] iceberg: If present and true, allocate fluxes for icebergs[in] salt: If present and true, allocate salt fluxes
-
subroutine, public mom_forcing_type::allocate_mech_forcing(G G, forces forces, stress stress, ustar ustar, shelf shelf, press press, iceberg iceberg) Conditionally allocate fields within the mechanical forcing type.
- Parameters
[in] g: Ocean grid structure[inout] forces: Forcing fields structure[in] stress: If present and true, allocate taux, tauy[in] ustar: If present and true, allocate ustar and related fields[in] shelf: If present and true, allocate forces for ice-shelf[in] press: If present and true, allocate p_surf and related fields[in] iceberg: If present and true, allocate forces for icebergs
-
subroutine
myalloc(array array, is is, ie ie, js js, je je, flag flag)¶ Allocates and zeroes-out array.
- Parameters
array: Array to be allocated[in] is: Start i-index[in] ie: End i-index[in] js: Start j-index[in] je: End j-index[in] flag: Flag to indicate to allocate
-
subroutine, public mom_forcing_type::deallocate_forcing_type(fluxes fluxes) Deallocate the forcing type.
- Parameters
[inout] fluxes: Forcing fields structure
-
subroutine, public mom_forcing_type::deallocate_mech_forcing(forces forces) Deallocate the mechanical forcing type.
- Parameters
[inout] forces: Forcing fields structure
-
-
namespace
mom_full_convection¶ Does full convective adjustment of unstable regions via a strong diffusivity.
Functions
-
subroutine, public mom_full_convection::full_convection(G G, GV GV, h h, tv tv, T_adj T_adj, S_adj S_adj, p_surf p_surf, Kddt_smooth Kddt_smooth, Kddt_convect Kddt_convect, halo halo) Calculate new temperatures and salinities that have been subject to full convective mixing.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][in] tv: A structure pointing to various thermodynamic variables[out] t_adj: Adjusted potential temperature [degC].[out] s_adj: Adjusted salinity [ppt].p_surf: The pressure at the ocean surface [Pa] (or NULL).[in] kddt_smooth: A smoothing vertical diffusivity times a timestep [H2 ~> m2 or kg2 m-4].[in] kddt_convect: A large convecting vertical diffusivity times a timestep [H2 ~> m2 or kg2 m-4].[in] halo: Halo width over which to compute
-
logical function mom_full_convection::is_unstable(dRho_dT dRho_dT, dRho_dS dRho_dS, h_a h_a, h_b h_b, mix_A mix_A, mix_B mix_B, T_a T_a, T_b T_b, S_a S_a, S_b S_b, Te_aa Te_aa, Te_bb Te_bb, Se_aa Se_aa, Se_bb Se_bb, d_A d_A, d_B d_B) This function returns True if the profiles around the given interface will be statically unstable after mixing above below. The arguments are the ocean state above and below, including partial calculations from a tridiagonal solver.
- Return
The return value, true if the profile is statically unstable around the interface in question.
- Parameters
[in] drho_dt: The derivative of in situ density with temperature [kg m-3 degC-1][in] drho_ds: The derivative of in situ density with salinity [kg m-3 ppt-1][in] h_a: The thickness of the layer above [H ~> m or kg m-2][in] h_b: The thickness of the layer below [H ~> m or kg m-2][in] mix_a: The time integrated mixing rate of the interface above [H ~> m or kg m-2][in] mix_b: The time integrated mixing rate of the interface below [H ~> m or kg m-2][in] t_a: The initial temperature of the layer above [degC][in] t_b: The initial temperature of the layer below [degC][in] s_a: The initial salinity of the layer below [ppt][in] s_b: The initial salinity of the layer below [ppt][in] te_aa: The estimated temperature two layers above rescaled by d_A [degC][in] te_bb: The estimated temperature two layers below rescaled by d_B [degC][in] se_aa: The estimated salinity two layers above rescaled by d_A [ppt][in] se_bb: The estimated salinity two layers below rescaled by d_B [ppt][in] d_a: The rescaling dependency across the interface above, nondim.[in] d_b: The rescaling dependency across the interface below, nondim.
-
subroutine
smoothed_drdt_drds(h h, tv tv, Kddt Kddt, dR_dT dR_dT, dR_dS dR_dS, G G, GV GV, j j, p_surf p_surf, halo halo)¶ Returns the partial derivatives of locally referenced potential density with temperature and salinity after the properties have been smoothed with a small constant diffusivity.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][in] tv: A structure pointing to various thermodynamic variables[in] kddt: A diffusivity times a time increment [H2 ~> m2 or kg2 m-4].[out] dr_dt: Derivative of locally referenced[out] dr_ds: Derivative of locally referenced[in] j: The j-point to work on.p_surf: The pressure at the ocean surface [Pa].[in] halo: Halo width over which to compute
-
-
namespace
mom_generic_tracer¶
-
namespace
mom_geothermal¶ Implemented geothermal heating at the ocean bottom.
Geothermal heating can be added either in a layered isopycnal mode, in which the heating raises the density of the layer to the target density of the layer above, and then moves the water into that layer, or in a simple Eulerian mode, in which the bottommost GEOTHERMAL_THICKNESS are heated. Geothermal heating will also provide a buoyant source of bottom TKE that can be used to further mix the near-bottom water. In cold fresh water lakes where heating increases density, water should be moved into deeper layers, but this is not implemented yet.
Functions
-
subroutine, public mom_geothermal::geothermal(h h, tv tv, dt dt, ea ea, eb eb, G G, GV GV, CS CS, halo halo) Applies geothermal heating, including the movement of water between isopycnal layers to match the target densities. The heating is applied to the bottommost layers that occur within ### of the bottom. If the partial derivative of the coordinate density with temperature is positive or very small, the layers are simply heated in place. Any heat that can not be applied to the ocean is returned (WHERE)?
- Parameters
[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[inout] h: Layer thicknesses [H ~> m or kg m-2][inout] tv: A structure containing pointers to any available thermodynamic fields. Absent fields have NULL ptrs.[in] dt: Time increment [s].[inout] ea: The amount of fluid moved downward into a layer; this should be increased due to mixed layer detrainment [H ~> m or kg m-2][inout] eb: The amount of fluid moved upward into a layer; this should be increased due to mixed layer entrainment [H ~> m or kg m-2].cs: The control structure returned by a previous call to geothermal_init.[in] halo: Halo width over which to work
-
subroutine, public mom_geothermal::geothermal_init(Time Time, G G, param_file param_file, diag diag, CS CS) Initialize parameters and allocate memory associated with the geothermal heating module.
- Parameters
[in] time: Current model time.[inout] g: The ocean’s grid structure.[in] param_file: A structure to parse for run-time parameters.[inout] diag: Structure used to regulate diagnostic output.cs: Pointer pointing to the module control structure.
-
subroutine, public mom_geothermal::geothermal_end(CS CS) Clean up and deallocate memory associated with the geothermal heating module.
- Parameters
cs: Geothermal heating control structure that will be deallocated in this subroutine.
-
-
namespace
mom_get_input¶ Reads the only Fortran name list needed to boot-strap the model.
The name list parameters indicate which directories to use for certain types of input and output, and which files to look in for the full parsable input parameter file(s).
Functions
-
subroutine, public mom_get_input::get_mom_input(param_file param_file, dirs dirs, check_params check_params, default_input_filename default_input_filename, ensemble_num ensemble_num) Get the names of the I/O directories and initialization file. Also calls the subroutine that opens run-time parameter files.
- Parameters
[out] param_file: A structure to parse for run-time parameters.[out] dirs: Container for paths and parameter file names.[in] check_params: If present and False will stop error checking for run-time parameters.[in] default_input_filename: If present, is the value assumed for input_filename if input_filename is not listed in the namelist MOM_input_nml.[in] ensemble_num: The ensemble id of the current member
-
-
namespace
mom_grid¶ Provides the ocean grid type.
Grid metrics and their inverses are labelled according to their staggered location on a Arakawa C (or B) grid.
Metrics centered on h- or T-points are labelled T, e.g. dxT is the distance across the cell in the x-direction.
Metrics centered on u-points are labelled Cu (C-grid u location). e.g. dyCu is the y-distance between two corners of a T-cell.
Metrics centered on v-points are labelled Cv (C-grid v location). e.g. dyCv is the y-distance between two -points.
Metrics centered on q-points are labelled Bu (B-grid u,v location). e.g. areaBu is the area centered on a q-point.
location on an T-cell and around a q-point.”
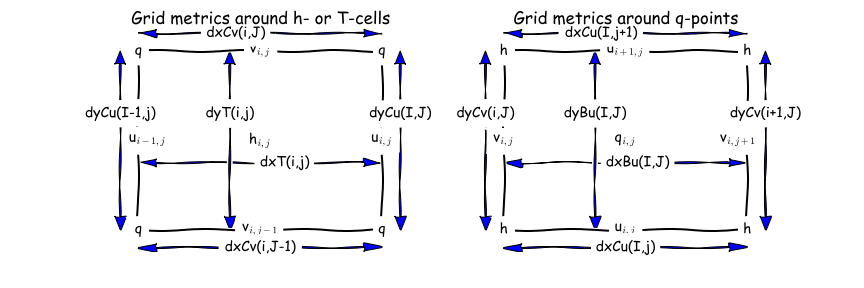
Areas centered at T-, u-, v- and q- points are
areaT,areaCu,areaCvandareaBurespectively.The reciprocal of metrics are pre-calculated and also stored in the ocean_grid_type with a I prepended to the name. For example,
1./areaTis calledIareaT, and1./dyCvisIdyCv.Geographic latitude and longitude (or model coordinates if not on a sphere) are stored in
geoLatT,geoLonTfor T-points. u-, v- and q- point coordinates are follow same pattern of replacing T with Cu, Cv and Bu respectively.Each location also has a 2D mask indicating whether the entire column is land or ocean.
mask2dTis 1 if the column is wet or 0 if the T-cell is land.mask2dCuis 1 if both neighboring column are ocean, and 0 if either is land.Functions
-
subroutine, public mom_grid::mom_grid_init(G G, param_file param_file, HI HI, global_indexing global_indexing, bathymetry_at_vel bathymetry_at_vel) MOM_grid_init initializes the ocean grid array sizes and grid memory.
- Parameters
[inout] g: The horizontal grid type[in] param_file: Parameter file handle[in] hi: A hor_index_type for array extents[in] global_indexing: If true use global index values instead of having the data domain on each processor start at 1.[in] bathymetry_at_vel: If true, there are separate values for the ocean bottom depths at velocity points. Otherwise the effects of topography are entirely determined from thickness points.
-
subroutine, public mom_grid::rescale_grid_bathymetry(G G, m_in_new_units m_in_new_units) rescale_grid_bathymetry permits a change in the internal units for the bathymetry on the grid, both rescaling the depths and recording the new internal units.
- Parameters
[inout] g: The horizontal grid structure[in] m_in_new_units: The new internal representation of 1 m depth.
-
subroutine, public mom_grid::set_derived_metrics(G G) set_derived_metrics calculates metric terms that are derived from other metrics.
- Parameters
[inout] g: The horizontal grid structure
-
real function mom_grid::adcroft_reciprocal(val val) Adcroft_reciprocal(x) = 1/x for |x|>0 or 0 for x=0.
- Return
The Adcroft reciprocal of val.
- Parameters
[in] val: The value being inverted.
-
logical function, public mom_grid::ispointincell(G G, i i, j j, x x, y y) Returns true if the coordinates (x,y) are within the h-cell (i,j)
- Parameters
[in] g: Grid type[in] i: i index of cell to test[in] j: j index of cell to test[in] x: x coordinate of point[in] y: y coordinate of point
-
subroutine, public mom_grid::set_first_direction(G G, y_first y_first) Store an integer indicating which direction to work on first.
- Parameters
[inout] g: The ocean’s grid structure[in] y_first: The first direction to store
-
subroutine, public mom_grid::get_global_grid_size(G G, niglobal niglobal, njglobal njglobal) Return global shape of horizontal grid.
- Parameters
[inout] g: The horizontal grid type[out] niglobal: i-index global size of grid[out] njglobal: j-index global size of grid
-
subroutine
allocate_metrics(G G)¶ Allocate memory used by the ocean_grid_type and related structures.
- Parameters
[inout] g: The horizontal grid type
-
subroutine, public mom_grid::mom_grid_end(G G) Release memory used by the ocean_grid_type and related structures.
- Parameters
[inout] g: The horizontal grid type
-
namespace
mom_grid_initialize¶ Initializes horizontal grid.
The metric terms have the form Dzp, IDzp, or DXDYp, where z can be X or Y, and p can be q, u, v, or h. z describes the direction of the metric, while p describes the location. IDzp is the inverse of Dzp, while DXDYp is the product of DXp and DYp except that areaT is calculated analytically from the latitudes and longitudes of the surrounding q points.
On a sphere, a variety of grids can be implemented by defining analytic expressions for dx_di, dy_dj (where x and y are latitude and longitude, and i and j are grid indices) and the expressions for the integrals of their inverses in the four subroutines dy_dj, Int_dj_dy, dx_di, and Int_di_dx.
initialize_masks sets up land masks based on the depth field. The one argument is the minimum ocean depth. Depths that are less than this are interpreted as land points.
Functions
-
subroutine, public mom_grid_initialize::set_grid_metrics(G G, param_file param_file, US US) set_grid_metrics is used to set the primary values in the model’s horizontal grid. The bathymetry, land-sea mask and any restricted channel widths are not known yet, so these are set later.
- Parameters
[inout] g: The dynamic horizontal grid type[in] param_file: Parameter file structure[in] us: A dimensional unit scaling type
-
subroutine
grid_metrics_chksum(parent parent, G G)¶ grid_metrics_chksum performs a set of checksums on metrics on the grid for debugging.
- Parameters
[in] parent: A string identifying the caller[in] g: The dynamic horizontal grid type
-
subroutine
set_grid_metrics_from_mosaic(G G, param_file param_file)¶ Sets the grid metrics from a mosaic file.
- Parameters
[inout] g: The dynamic horizontal grid type[in] param_file: Parameter file structure
-
subroutine
set_grid_metrics_cartesian(G G, param_file param_file)¶ Calculate the values of the metric terms for a Cartesian grid that might be used and save them in arrays.
Within this subroutine, the x- and y- grid spacings and their inverses and the cell areas centered on h, q, u, and v points are calculated, as are the geographic locations of each of these 4 sets of points.
- Parameters
[inout] g: The dynamic horizontal grid type[in] param_file: Parameter file structure
-
subroutine
set_grid_metrics_spherical(G G, param_file param_file)¶ Calculate the values of the metric terms that might be used and save them in arrays.
Within this subroutine, the x- and y- grid spacings and their inverses and the cell areas centered on h, q, u, and v points are calculated, as are the geographic locations of each of these 4 sets of points.
- Parameters
[inout] g: The dynamic horizontal grid type[in] param_file: Parameter file structure
-
subroutine
set_grid_metrics_mercator(G G, param_file param_file)¶ Calculate the values of the metric terms that might be used and save them in arrays.
Within this subroutine, the x- and y- grid spacings and their inverses and the cell areas centered on h, q, u, and v points are calculated, as are the geographic locations of each of these 4 sets of points.
- Parameters
[inout] g: The dynamic horizontal grid type[in] param_file: Parameter file structure
-
real function mom_grid_initialize::ds_di(x x, y y, GP GP) This function returns the grid spacing in the logical x direction.
- Parameters
[in] x: The longitude in question[in] y: The latitude in question[in] gp: A structure of grid parameters
-
real function mom_grid_initialize::ds_dj(x x, y y, GP GP) This function returns the grid spacing in the logical y direction.
- Parameters
[in] x: The longitude in question[in] y: The latitude in question[in] gp: A structure of grid parameters
-
real function mom_grid_initialize::dl(x1 x1, x2 x2, y1 y1, y2 y2) This function returns the contribution from the line integral along one of the four sides of a cell face to the area of a cell, assuming that the sides follow a linear path in latitude and longitude (i.e., on a Mercator grid).
- Parameters
[in] x1: Segment starting longitude, in degrees E.[in] x2: Segment ending longitude, in degrees E.[in] y1: Segment ending latitude, in degrees N.[in] y2: Segment ending latitude, in degrees N.
-
real function mom_grid_initialize::find_root(fn fn, dy_df dy_df, GP GP, fnval fnval, y1 y1, ymin ymin, ymax ymax, ittmax ittmax) This subroutine finds and returns the value of y at which the monotonically increasing function fn takes the value fnval, also returning in ittmax the number of iterations of Newton’s method that were used to polish the root.
- Return
The value of y where fn(y) = fnval that will be returned
- Parameters
fn: The external function whose root is being soughtdy_df: The inverse of the derivative of that function[in] gp: A structure of grid parameters[in] fnval: The value of fn being sought[in] y1: A first guess for y[in] ymin: The minimum permitted value of y[in] ymax: The maximum permitted value of y[out] ittmax: The number of iterations used to polish the root
-
real function mom_grid_initialize::dx_di(x x, GP GP) This function calculates and returns the value of dx/di, where x is the longitude in Radians, and i is the integral north-south grid index.
- Parameters
[in] x: The longitude in question[in] gp: A structure of grid parameters
-
real function mom_grid_initialize::int_di_dx(x x, GP GP) This function calculates and returns the integral of the inverse of dx/di to the point x, in radians.
- Parameters
[in] x: The longitude in question[in] gp: A structure of grid parameters
-
real function mom_grid_initialize::dy_dj(y y, GP GP) This subroutine calculates and returns the value of dy/dj, where y is the latitude in Radians, and j is the integral north-south grid index.
- Parameters
[in] y: The latitude in question[in] gp: A structure of grid parameters
-
real function mom_grid_initialize::int_dj_dy(y y, GP GP) This subroutine calculates and returns the integral of the inverse of dy/dj to the point y, in radians.
- Parameters
[in] y: The latitude in question[in] gp: A structure of grid parameters
-
subroutine
extrapolate_metric(var var, jh jh, missing missing)¶ Extrapolates missing metric data into all the halo regions.
- Parameters
[inout] var: The array in which to fill in halos[in] jh: The size of the halos to be filled[in] missing: The missing data fill value, 0 by default.
-
real function, public mom_grid_initialize::adcroft_reciprocal(val val) This function implements Adcroft’s rule for reciprocals, namely that Adcroft_Inv(x) = 1/x for |x|>0 or 0 for x=0.
- Return
The Adcroft reciprocal of val.
- Parameters
[in] val: The value being inverted.
-
subroutine, public mom_grid_initialize::initialize_masks(G G, PF PF, US US) Initializes the grid masks and any metrics that come with masks already applied.
Initialize_masks sets mask2dT, mask2dCu, mask2dCv, and mask2dBu to mask out flow over any points which are shallower than Dmin and permit an appropriate treatment of the boundary conditions. mask2dCu and mask2dCv are 0.0 at any points adjacent to a land point. mask2dBu is 0.0 at any land or boundary point. For points in the interior, mask2dCu, mask2dCv, and mask2dBu are all 1.0.
- Parameters
[inout] g: The dynamic horizontal grid type[in] pf: Parameter file structure[in] us: A dimensional unit scaling type
-
-
namespace
mom_hor_index¶ Defines the horizontal index type (hor_index_type) used for providing index ranges.
The hor_index_type provides the decalarations and loop ranges for almost all data with horizontal extent.
Declarations and loop ranges should always be coded with the symmetric memory model in mind. The non-symmetric memory mode will then also work, albeit with a different (less efficient) communication pattern.
Using the hor_index_type HI:
declaration of h-point data is of the form
h(HI%isd:HI%ied,HI%jsd:HI%jed)declaration of q-point data is of the form
q(HI%IsdB:HI%IedB,HI%JsdB:HI%JedB)declaration of u-point data is of the form
u(HI%IsdB:HI%IedB,HI%jsd:HI%jed)declaration of v-point data is of the form
v(HI%isd:HI%ied,HI%JsdB:HI%JedB).
For more detail explanation of horizontal indexing see Horizontal indexing and memory.
Functions
-
subroutine, public mom_hor_index::hor_index_init(Domain Domain, HI HI, param_file param_file, local_indexing local_indexing, index_offset index_offset) Sets various index values in a hor_index_type.
- Parameters
[in] domain: The MOM domain from which to extract information.[inout] hi: A horizontal index type to populate with data[in] param_file: Parameter file handle[in] local_indexing: If true, all tracer data domains start at 1[in] index_offset: A fixed additional offset to all indices
-
subroutine
hit_assign(HI1 HI1, HI2 HI2)¶ HIT_assign copies one hor_index_type into another. It is accessed via an assignment (=) operator.
- Parameters
[out] hi1: Horizontal index type to copy to[in] hi2: Horizontal index type to copy from
-
namespace
mom_hor_visc¶ Calculates horizontal viscosity and viscous stresses.
This module contains the subroutine horizontal_viscosity() that calculates the effects of horizontal viscosity, including parameterizations of the value of the viscosity itself. horizontal_viscosity() calculates the acceleration due to some combination of a biharmonic viscosity and a Laplacian viscosity. Either or both may use a coefficient that depends on the shear and strain of the flow. All metric terms are retained. The Laplacian is calculated as the divergence of a stress tensor, using the form suggested by Smagorinsky (1993). The biharmonic is calculated by twice applying the divergence of the stress tensor that is used to calculate the Laplacian, but without the dependence on thickness in the first pass. This form permits a variable viscosity, and indicates no acceleration for either resting fluid or solid body rotation.
The form of the viscous accelerations is discussed extensively in Griffies and Hallberg (2000), and the implementation here follows that discussion closely. We use the notation of Smith and McWilliams (2003) with the exception that the isotropic viscosity is \(\kappa_h\).
Unnamed Group
-
subroutine, public mom_hor_visc::horizontal_viscosity(u u, v v, h h, diffu diffu, diffv diffv, MEKE MEKE, VarMix VarMix, G G, GV GV, US US, CS CS, OBC OBC, BT BT) Calculates the acceleration due to the horizontal viscosity.
A combination of biharmonic and Laplacian forms can be used. The coefficient may either be a constant or a shear-dependent form. The biharmonic is determined by twice taking the divergence of an appropriately defined stress tensor. The Laplacian is determined by doing so once.
To work, the following fields must be set outside of the usual is:ie range before this subroutine is called: u[is-2:ie+2,js-2:je+2] v[is-2:ie+2,js-2:je+2] h[is-1:ie+1,js-1:je+1]
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] u: The zonal velocity [m s-1].[in] v: The meridional velocity [m s-1].[inout] h: Layer thicknesses [H ~> m or kg m-2].[out] diffu: Zonal acceleration due to convergence of[out] diffv: Meridional acceleration due to convergencemeke: Pointer to a structure containing fields related to Mesoscale Eddy Kinetic Energy.varmix: Pointer to a structure with fields that specify the spatially variable viscosities[in] us: A dimensional unit scaling typecs: Control structure returned by a previous call to hor_visc_init.obc: Pointer to an open boundary condition typebt: Pointer to a structure containing barotropic velocities.
-
subroutine, public mom_hor_visc::hor_visc_init(Time Time, G G, US US, param_file param_file, diag diag, CS CS, MEKE MEKE) Allocates space for and calculates static variables used by horizontal_viscosity(). hor_visc_init calculates and stores the values of a number of metric functions that are used in horizontal_viscosity().
- Parameters
[in] time: Current model time.[inout] g: The ocean’s grid structure.[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters.[inout] diag: Structure to regulate diagnostic output.cs: Pointer to the control structure for this modulemeke: MEKE data
-
subroutine
align_aniso_tensor_to_grid(CS CS, n1 n1, n2 n2)¶ Calculates factors in the anisotropic orientation tensor to be align with the grid. With n1=1 and n2=0, this recovers the approach of Large et al, 2001.
- Parameters
cs: Control structure for horizontal viscosity[in] n1: i-component of direction vector [nondim][in] n2: j-component of direction vector [nondim]
-
subroutine
smooth_gme(CS CS, G G, GME_flux_h GME_flux_h, GME_flux_q GME_flux_q)¶ Apply a 1-1-4-1-1 Laplacian filter one time on GME diffusive flux to reduce any horizontal two-grid-point noise.
- Parameters
cs: Control structure[in] g: Ocean grid[inout] gme_flux_h: GME diffusive flux at h points[inout] gme_flux_q: GME diffusive flux at q points
-
subroutine, public mom_hor_visc::hor_visc_end(CS CS) Deallocates any variables allocated in hor_visc_init.
- Parameters
cs: The control structure returned by a previous call to hor_visc_init.
-
-
namespace
mom_horizontal_regridding¶ Horizontal interpolation.
Unnamed Group
-
subroutine, public mom_horizontal_regridding::mystats(array array, missing missing, is is, ie ie, js js, je je, k k, mesg mesg) Write to the terminal some basic statistics about the k-th level of an array.
- Parameters
[in] array: input array (ND)[in] missing: missing value (ND)is: Horizontal loop bounds to calculate statistics forie: Horizontal loop bounds to calculate statistics forjs: Horizontal loop bounds to calculate statistics forje: Horizontal loop bounds to calculate statistics fork: Level to calculate statistics formesg: Label to use in message
-
subroutine
horiz_interp_and_extrap_tracer_fms_id(fms_id fms_id, Time Time, conversion conversion, G G, tr_z tr_z, mask_z mask_z, z_in z_in, z_edges_in z_edges_in, missing_value missing_value, reentrant_x reentrant_x, tripolar_n tripolar_n, homogenize homogenize, m_to_Z m_to_Z)¶ Extrapolate and interpolate using a FMS time interpolation handle.
- Parameters
[in] fms_id: A unique id used by the FMS time interpolator[in] time: A FMS time type[in] conversion: Conversion factor for tracer.[inout] g: Grid objecttr_z: pointer to allocatable tracer array on local model grid and native vertical levels.mask_z: pointer to allocatable tracer mask array on local model grid and native vertical levels.z_in: Cell grid values for input data.z_edges_in: Cell grid edge values for input data. (Intent out)[out] missing_value: The missing value in the returned array.[in] reentrant_x: If true, this grid is reentrant in the x-direction[in] tripolar_n: If true, this is a northern tripolar grid[in] homogenize: If present and true, horizontally homogenize data to produce perfectly “flat” initial conditions[in] m_to_z: A conversion factor from meters to the units of depth. If missing, GbathyT must be in m.
-
subroutine
meshgrid(x x, y y, x_T x_T, y_T y_T)¶ Create a 2d-mesh of grid coordinates from 1-d arrays.
- Parameters
[in] x: input 1-dimensional vector[in] y: input 1-dimensional vector[inout] x_t: output 2-dimensional array[inout] y_t: output 2-dimensional array
-
integer function, dimension(0:size(m,1)+1,0:size(m,2)+1) mom_horizontal_regridding::fill_boundaries_int(m m, cyclic_x cyclic_x, tripolar_n tripolar_n) Fill grid edges for integer data.
- Parameters
[in] m: input array (ND)[in] cyclic_x: True if domain is zonally re-entrant[in] tripolar_n: True if domain has an Arctic fold
-
real function, dimension(0:size(m,1)+1,0:size(m,2)+1) mom_horizontal_regridding::fill_boundaries_real(m m, cyclic_x cyclic_x, tripolar_n tripolar_n) Fill grid edges for real data.
- Parameters
[in] m: input array (ND)[in] cyclic_x: True if domain is zonally re-entrant[in] tripolar_n: True if domain has an Arctic fold
-
subroutine
smooth_heights(zi zi, fill fill, bad bad, sor sor, niter niter, cyclic_x cyclic_x, tripolar_n tripolar_n)¶ Solve del2 (zi) = 0 using successive iterations with a 5 point stencil. Only points fill==1 are modified. Except where bad==1, information propagates isotropically in index space. The resulting solution in each region is an approximation to del2(zi)=0 subject to boundary conditions along the valid points curve bounding this region.
- Parameters
[inout] zi: input and output array (ND)[in] fill: same shape as zi, 1=fill[in] bad: same shape as zi, 1=bad data[in] sor: relaxation coefficient (ND)[in] niter: maximum number of iterations[in] cyclic_x: true if domain is zonally reentrant[in] tripolar_n: true if domain has an Arctic fold
Functions
-
subroutine
fill_miss_2d(aout aout, good good, fill fill, prev prev, G G, smooth smooth, num_pass num_pass, relc relc, crit crit, keep_bug keep_bug, debug debug)¶ Use ICE-9 algorithm to populate points (fill=1) with valid data (good=1). If no information is available, Then use a previous guess (prev). Optionally (smooth) blend the filled points to achieve a more desirable result.
- Parameters
[inout] g: The ocean’s grid structure.[inout] aout: The array with missing values to fill[in] good: Valid data mask for incoming array[in] fill: Same shape array of points which need[in] prev: First guess where isolated holes exist.[in] smooth: If present and true, apply a number of Laplacian iterations to the interpolated data[in] num_pass: The maximum number of iterations[in] relc: A relaxation coefficient for Laplacian (ND)[in] crit: A minimal value for deltas between iterations.[in] keep_bug: Use an algorithm with a bug that dates to the “sienna” code release.[in] debug: If true, write verbose debugging messages.
-
subroutine
horiz_interp_and_extrap_tracer_record(filename filename, varnam varnam, conversion conversion, recnum recnum, G G, tr_z tr_z, mask_z mask_z, z_in z_in, z_edges_in z_edges_in, missing_value missing_value, reentrant_x reentrant_x, tripolar_n tripolar_n, homogenize homogenize, m_to_Z m_to_Z)¶ Extrapolate and interpolate from a file record.
- Parameters
[in] filename: Path to file containing tracer to be interpolated.[in] varnam: Name of tracer in filee.[in] conversion: Conversion factor for tracer.[in] recnum: Record number of tracer to be read.[inout] g: Grid objecttr_z: pointer to allocatable tracer array on local model grid and input-file vertical levels.mask_z: pointer to allocatable tracer mask array on local model grid and input-file vertical levels.z_in: Cell grid values for input data.z_edges_in: Cell grid edge values for input data.[out] missing_value: The missing value in the returned array.[in] reentrant_x: If true, this grid is reentrant in the x-direction[in] tripolar_n: If true, this is a northern tripolar grid[in] homogenize: If present and true, horizontally homogenize data to produce perfectly “flat” initial conditions[in] m_to_z: A conversion factor from meters to the units of depth. If missing, GbathyT must be in m.
-
-
namespace
mom_ice_shelf¶ Implements the thermodynamic aspects of ocean / ice-shelf interactions, along with a crude placeholder for a later implementation of full ice shelf dynamics, all using the MOM framework and coding style.
Functions
-
subroutine, public mom_ice_shelf::shelf_calc_flux(state state, fluxes fluxes, Time Time, time_step time_step, CS CS, forces forces) Calculates fluxes between the ocean and ice-shelf using the three-equations formulation (optional to use just two equations). See ICE_SHELF equations.
- Parameters
[inout] state: structure containing fields that describe the surface state of the ocean[inout] fluxes: structure containing pointers to any possible thermodynamic or mass-flux forcing fields.[in] time: Start time of the fluxes.[in] time_step: Length of time over which these fluxes will be applied [s].cs: A pointer to the control structure returned by a previous call to initialize_ice_shelf.[inout] forces: A structure with the driving mechanical forces
-
subroutine
change_thickness_using_melt(ISS ISS, G G, time_step time_step, fluxes fluxes, rho_ice rho_ice, debug debug)¶ Changes the thickness (mass) of the ice shelf based on sub-ice-shelf melting.
- Parameters
[inout] g: The ocean’s grid structure.[inout] iss: A structure with elements that describe the ice-shelf state[in] time_step: The time step for this update [s].[inout] fluxes: structure containing pointers to any possible thermodynamic or mass-flux forcing fields.[in] rho_ice: The density of ice-shelf ice [kg m-2 Z-1 ~> kg m-3].[in] debug: If present and true, write chksums
-
subroutine, public mom_ice_shelf::add_shelf_forces(G G, CS CS, forces forces, do_shelf_area do_shelf_area) This subroutine adds the mechanical forcing fields and perhaps shelf areas, based on the ice state in ice_shelf_CS.
- Parameters
[inout] g: The ocean’s grid structure.cs: This module’s control structure.[inout] forces: A structure with the driving mechanical forces[in] do_shelf_area: If true find the shelf-covered areas.
-
subroutine
add_shelf_pressure(G G, CS CS, fluxes fluxes)¶ This subroutine adds the ice shelf pressure to the fluxes type.
- Parameters
[inout] g: The ocean’s grid structure.[in] cs: This module’s control structure.[inout] fluxes: A structure of surface fluxes that may be updated.
-
subroutine, public mom_ice_shelf::add_shelf_flux(G G, CS CS, state state, fluxes fluxes) Updates surface fluxes that are influenced by sub-ice-shelf melting.
- Parameters
[inout] g: The ocean’s grid structure.cs: This module’s control structure.[inout] state: Surface ocean state[inout] fluxes: A structure of surface fluxes that may be used/updated.
-
subroutine, public mom_ice_shelf::initialize_ice_shelf(param_file param_file, ocn_grid ocn_grid, Time Time, CS CS, diag diag, forces forces, fluxes fluxes, Time_in Time_in, solo_ice_sheet_in solo_ice_sheet_in) Initializes shelf model data, parameters and diagnostics.
- Parameters
[in] param_file: A structure to parse for run-time parametersocn_grid: The calling ocean model’s horizontal grid structure[inout] time: The clock that that will indicate the model timecs: A pointer to the ice shelf control structure[in] diag: A structure that is used to regulate the diagnostic output.[inout] fluxes: A structure containing pointers to any possible thermodynamic or mass-flux forcing fields.[inout] forces: A structure with the driving mechanical forces[in] time_in: The time at initialization.[in] solo_ice_sheet_in: If present, this indicates whether a solo ice-sheet driver.
-
subroutine
initialize_shelf_mass(G G, param_file param_file, CS CS, ISS ISS, new_sim new_sim)¶ Initializes shelf mass based on three options (file, zero and user)
- Parameters
[in] g: The ocean’s grid structure.[in] param_file: A structure to parse for run-time parameterscs: A pointer to the ice shelf control structure[inout] iss: The ice shelf state type that is being updated[in] new_sim: If present and false, this run is being restarted
-
subroutine
update_shelf_mass(G G, CS CS, ISS ISS, Time Time)¶ Updates the ice shelf mass using data from a file.
- Parameters
[inout] g: The ocean’s grid structure.[in] cs: A pointer to the ice shelf control structure[inout] iss: The ice shelf state type that is being updated[in] time: The current model time
-
subroutine, public mom_ice_shelf::ice_shelf_save_restart(CS CS, Time Time, directory directory, time_stamped time_stamped, filename_suffix filename_suffix) Save the ice shelf restart file.
- Parameters
cs: ice shelf control structure[in] time: model time at this call[in] directory: An optional directory into which to write these restart files.[in] time_stamped: f true, the restart file names include a unique time stamp. The default is false.[in] filename_suffix: An optional suffix (e.g., a time-stamp) to append to the restart file names.
-
subroutine, public mom_ice_shelf::ice_shelf_end(CS CS) Deallocates all memory associated with this module.
- Parameters
cs: A pointer to the ice shelf control structure
-
subroutine, public mom_ice_shelf::solo_time_step(CS CS, time_step time_step, nsteps nsteps, Time Time, min_time_step_in min_time_step_in) This routine is for stepping a stand-alone ice shelf model without an ocean.
- Parameters
cs: A pointer to the ice shelf control structure[in] time_step: The time interval for this update [s].[inout] nsteps: The running number of ice shelf steps.[inout] time: The current model time[in] min_time_step_in: The minimum permitted time step [s].
-
-
namespace
mom_ice_shelf_dynamics¶ Implements a crude placeholder for a later implementation of full ice shelf dynamics.
Unnamed Group
-
real function mom_ice_shelf_dynamics::slope_limiter(num num, denom denom) used for flux limiting in advective subroutines Van Leer limiter (source: Wikipedia)
- Parameters
[in] num: The numerator of the ratio used in the Van Leer slope limiter[in] denom: The denominator of the ratio used in the Van Leer slope limiter
-
real function mom_ice_shelf_dynamics::quad_area(X X, Y Y) Calculate area of quadrilateral.
- Parameters
[in] x: The x-positions of the vertices of the quadrilateral.[in] y: The y-positions of the vertices of the quadrilateral.
-
subroutine, public mom_ice_shelf_dynamics::register_ice_shelf_dyn_restarts(G G, param_file param_file, CS CS, restart_CS restart_CS) This subroutine is used to register any fields related to the ice shelf dynamics that should be written to or read from the restart file.
- Parameters
[inout] g: The grid type describing the ice shelf grid.[in] param_file: A structure to parse for run-time parameterscs: A pointer to the ice shelf dynamics control structurerestart_cs: A pointer to the restart control structure.
-
subroutine, public mom_ice_shelf_dynamics::initialize_ice_shelf_dyn(param_file param_file, Time Time, ISS ISS, CS CS, G G, US US, diag diag, new_sim new_sim, solo_ice_sheet_in solo_ice_sheet_in) Initializes shelf model data, parameters and diagnostics.
- Parameters
[in] param_file: A structure to parse for run-time parameters[inout] time: The clock that that will indicate the model time[in] iss: A structure with elements that describe the ice-shelf statecs: A pointer to the ice shelf dynamics control structure[inout] g: The grid type describing the ice shelf grid.[in] us: Pointer to a structure containing unit conversion factors[in] diag: A structure that is used to regulate the diagnostic output.[in] new_sim: If true this is a new simulation, otherwise has been started from a restart file.[in] solo_ice_sheet_in: If present, this indicates whether a solo ice-sheet driver.
-
subroutine
initialize_diagnostic_fields(CS CS, ISS ISS, G G, US US, Time Time)¶ - Parameters
[inout] cs: A pointer to the ice shelf control structure[in] iss: A structure with elements that describe the ice-shelf state[inout] g: The grid structure used by the ice shelf.[in] us: Pointer to a structure containing unit conversion factors[in] time: The current model time
-
real function, public mom_ice_shelf_dynamics::ice_time_step_cfl(CS CS, ISS ISS, G G) This function returns the global maximum timestep that can be taken based on the current ice velocities. Because it involves finding a global minimum, it can be suprisingly expensive.
- Return
The maximum permitted timestep based on the ice velocities [s].
- Parameters
[inout] cs: The ice shelf dynamics control structure[inout] iss: A structure with elements that describe the ice-shelf state[inout] g: The grid structure used by the ice shelf.
-
subroutine, public mom_ice_shelf_dynamics::update_ice_shelf(CS CS, ISS ISS, G G, US US, time_step time_step, Time Time, ocean_mass ocean_mass, coupled_grounding coupled_grounding, must_update_vel must_update_vel) This subroutine updates the ice shelf velocities, mass, stresses and properties due to the ice shelf dynamics.
- Parameters
[inout] cs: The ice shelf dynamics control structure[inout] iss: A structure with elements that describe the ice-shelf state[inout] g: The grid structure used by the ice shelf.[in] us: Pointer to a structure containing unit conversion factors[in] time_step: time step [s][in] time: The current model time[in] ocean_mass: If present this is the mass per unit area[in] coupled_grounding: If true, the grounding line is determined by coupled ice-ocean dynamics[in] must_update_vel: Always update the ice velocities if true.
-
subroutine
ice_shelf_advect(CS CS, ISS ISS, G G, time_step time_step, Time Time)¶ This subroutine takes the velocity (on the Bgrid) and timesteps h_t = - div (uh) once. Additionally, it will update the volume of ice in partially-filled cells, and update hmask accordingly.
- Parameters
[inout] cs: The ice shelf dynamics control structure[inout] iss: A structure with elements that describe the ice-shelf state[inout] g: The grid structure used by the ice shelf.[in] time_step: time step [s][in] time: The current model time
-
subroutine
ice_shelf_solve_outer(CS CS, ISS ISS, G G, US US, u u, v v, iters iters, time time)¶ - Parameters
[inout] cs: The ice shelf dynamics control structure[in] iss: A structure with elements that describe the ice-shelf state[inout] g: The grid structure used by the ice shelf.[in] us: Pointer to a structure containing unit conversion factors[inout] u: The zonal ice shelf velocity at vertices [m year-1][inout] v: The meridional ice shelf velocity at vertices [m year-1][out] iters: The number of iterations used in the solver.[in] time: The current model time
-
subroutine
ice_shelf_solve_inner(CS CS, ISS ISS, G G, u u, v v, taudx taudx, taudy taudy, H_node H_node, float_cond float_cond, hmask hmask, conv_flag conv_flag, iters iters, time time, Phi Phi, Phisub Phisub)¶ - Parameters
[in] cs: A pointer to the ice shelf control structure[in] iss: A structure with elements that describe the ice-shelf state[inout] g: The grid structure used by the ice shelf.[inout] u: The zonal ice shelf velocity at vertices [m year-1][inout] v: The meridional ice shelf velocity at vertices [m year-1][in] taudx: The x-direction driving stress, in ???[in] taudy: The y-direction driving stress, in ???[in] h_node: The ice shelf thickness at nodal (corner)[in] float_cond: An array indicating where the ice[in] hmask: A mask indicating which tracer points are[out] conv_flag: A flag indicating whether (1) or not (0) the iterations have converged to the specified tolerence[out] iters: The number of iterations used in the solver.[in] time: The current model time[in] phi: The gradients of bilinear basis elements at Gaussian[in] phisub: Quadrature structure weights at subgridscale
-
subroutine
ice_shelf_advect_thickness_x(CS CS, G G, time_step time_step, hmask hmask, h0 h0, h_after_uflux h_after_uflux, flux_enter flux_enter)¶ - Parameters
[in] cs: A pointer to the ice shelf control structure[in] g: The grid structure used by the ice shelf.[in] time_step: The time step for this update [s].[inout] hmask: A mask indicating which tracer points are[in] h0: The initial ice shelf thicknesses [Z ~> m].[inout] h_after_uflux: The ice shelf thicknesses after[inout] flux_enter: The ice volume flux into the cell
-
subroutine
ice_shelf_advect_thickness_y(CS CS, G G, time_step time_step, hmask hmask, h_after_uflux h_after_uflux, h_after_vflux h_after_vflux, flux_enter flux_enter)¶ - Parameters
[in] cs: A pointer to the ice shelf control structure[in] g: The grid structure used by the ice shelf.[in] time_step: The time step for this update [s].[inout] hmask: A mask indicating which tracer points are[in] h_after_uflux: The ice shelf thicknesses after[inout] h_after_vflux: The ice shelf thicknesses after[inout] flux_enter: The ice volume flux into the cell
-
subroutine, public mom_ice_shelf_dynamics::shelf_advance_front(CS CS, ISS ISS, G G, flux_enter flux_enter) - Parameters
[in] cs: A pointer to the ice shelf control structure[inout] iss: A structure with elements that describe the ice-shelf state[in] g: The grid structure used by the ice shelf.[inout] flux_enter: The ice volume flux into the cell
-
subroutine, public mom_ice_shelf_dynamics::ice_shelf_min_thickness_calve(G G, h_shelf h_shelf, area_shelf_h area_shelf_h, hmask hmask, thickness_calve thickness_calve) Apply a very simple calving law using a minimum thickness rule.
- Parameters
[in] g: The grid structure used by the ice shelf.[inout] h_shelf: The ice shelf thickness [Z ~> m].[inout] area_shelf_h: The area per cell covered by the ice shelf [m2].[inout] hmask: A mask indicating which tracer points are[in] thickness_calve: The thickness at which to trigger calving [Z ~> m].
-
subroutine, public mom_ice_shelf_dynamics::calve_to_mask(G G, h_shelf h_shelf, area_shelf_h area_shelf_h, hmask hmask, calve_mask calve_mask) - Parameters
[in] g: The grid structure used by the ice shelf.[inout] h_shelf: The ice shelf thickness [Z ~> m].[inout] area_shelf_h: The area per cell covered by the ice shelf [m2].[inout] hmask: A mask indicating which tracer points are[in] calve_mask: A mask that indicates where the ice shelf
-
subroutine
calc_shelf_driving_stress(CS CS, ISS ISS, G G, US US, TAUD_X TAUD_X, TAUD_Y TAUD_Y, OD OD)¶ - Parameters
[in] cs: A pointer to the ice shelf control structure[in] iss: A structure with elements that describe the ice-shelf state[inout] g: The grid structure used by the ice shelf.[in] us: Pointer to a structure containing unit conversion factors[in] od: ocean floor depth at tracer points [Z ~> m].[inout] taud_x: X-direction driving stress at q-points[inout] taud_y: Y-direction driving stress at q-points
-
subroutine
init_boundary_values(CS CS, G G, time time, hmask hmask, input_flux input_flux, input_thick input_thick, new_sim new_sim)¶ - Parameters
[inout] cs: A pointer to the ice shelf control structure[inout] g: The grid structure used by the ice shelf.[in] time: The current model time[in] hmask: A mask indicating which tracer points are[in] input_flux: The integrated inward ice thickness flux [Z m2 s-1 ~> m3 s-1][in] input_thick: The ice thickness at boundaries [Z ~> m].[in] new_sim: If present and false, this run is being restarted
-
subroutine
cg_action(uret uret, vret vret, u u, v v, Phi Phi, Phisub Phisub, umask umask, vmask vmask, hmask hmask, H_node H_node, nu nu, float_cond float_cond, bathyT bathyT, beta beta, dxdyh dxdyh, G G, is is, ie ie, js js, je je, dens_ratio dens_ratio)¶ - Parameters
[in] g: The grid structure used by the ice shelf.[inout] uret: The retarding stresses working at u-points.[inout] vret: The retarding stresses working at v-points.[in] phi: The gradients of bilinear basis elements at Gaussian[in] phisub: Quadrature structure weights at subgridscale[in] u: The zonal ice shelf velocity at vertices [m year-1][in] v: The meridional ice shelf velocity at vertices [m year-1][in] umask: A coded mask indicating the nature of the[in] vmask: A coded mask indicating the nature of the[in] h_node: The ice shelf thickness at nodal (corner)[in] hmask: A mask indicating which tracer points are[in] nu: A field related to the ice viscosity from Glen’s[in] float_cond: An array indicating where the ice[in] bathyt: The depth of ocean bathymetry at tracer points [Z ~> m].[in] beta: A field related to the nonlinear part of the[in] dxdyh: The tracer cell area [m2][in] dens_ratio: The density of ice divided by the density of seawater, nondimensional[in] is: The starting i-index to work on[in] ie: The ending i-index to work on[in] js: The starting j-index to work on[in] je: The ending j-index to work on
-
subroutine
cg_action_subgrid_basal(Phisub Phisub, H H, U U, V V, DXDYH DXDYH, bathyT bathyT, dens_ratio dens_ratio, Ucontr Ucontr, Vcontr Vcontr)¶ - Parameters
[in] phisub: Quadrature structure weights at subgridscale[in] h: The ice shelf thickness at nodal (corner) points [Z ~> m].[in] u: The zonal ice shelf velocity at vertices [m year-1][in] v: The meridional ice shelf velocity at vertices [m year-1][in] dxdyh: The tracer cell area [m2][in] bathyt: The depth of ocean bathymetry at tracer points [Z ~> m].[in] dens_ratio: The density of ice divided by the density of seawater, nondimensional[inout] ucontr: A field related to the subgridscale contributions to the u-direction basal stress.[inout] vcontr: A field related to the subgridscale contributions to the v-direction basal stress.
-
subroutine
matrix_diagonal(CS CS, G G, float_cond float_cond, H_node H_node, nu nu, beta beta, hmask hmask, dens_ratio dens_ratio, Phisub Phisub, u_diagonal u_diagonal, v_diagonal v_diagonal)¶ returns the diagonal entries of the matrix for a Jacobi preconditioning
- Parameters
[in] cs: A pointer to the ice shelf control structure[in] g: The grid structure used by the ice shelf.[in] float_cond: An array indicating where the ice[in] h_node: The ice shelf thickness at nodal[in] nu: A field related to the ice viscosity from Glen’s[in] beta: A field related to the nonlinear part of the[in] hmask: A mask indicating which tracer points are[in] dens_ratio: The density of ice divided by the density of seawater, nondimensional[in] phisub: Quadrature structure weights at subgridscale locations for finite element calculations[inout] u_diagonal: The diagonal elements of the u-velocity[inout] v_diagonal: The diagonal elements of the v-velocity
-
subroutine
cg_diagonal_subgrid_basal(Phisub Phisub, H_node H_node, DXDYH DXDYH, bathyT bathyT, dens_ratio dens_ratio, Ucontr Ucontr, Vcontr Vcontr)¶ - Parameters
[in] phisub: Quadrature structure weights at subgridscale[in] h_node: The ice shelf thickness at nodal (corner) points [Z ~> m].[in] dxdyh: The tracer cell area [m2][in] bathyt: The depth of ocean bathymetry at tracer points [Z ~> m].[in] dens_ratio: The density of ice divided by the density of seawater, nondimensional[inout] ucontr: A field related to the subgridscale contributions to the u-direction diagonal elements from basal stress.[inout] vcontr: A field related to the subgridscale contributions to the v-direction diagonal elements from basal stress.
-
subroutine
apply_boundary_values(CS CS, ISS ISS, G G, time time, Phisub Phisub, H_node H_node, nu nu, beta beta, float_cond float_cond, dens_ratio dens_ratio, u_bdry_contr u_bdry_contr, v_bdry_contr v_bdry_contr)¶ - Parameters
[in] cs: A pointer to the ice shelf control structure[in] iss: A structure with elements that describe the ice-shelf state[in] g: The grid structure used by the ice shelf.[in] time: The current model time[in] phisub: Quadrature structure weights at subgridscale[in] h_node: The ice shelf thickness at nodal[in] nu: A field related to the ice viscosity from Glen’s[in] beta: A field related to the nonlinear part of the[in] float_cond: An array indicating where the ice[in] dens_ratio: The density of ice divided by the density of seawater, nondimensional[inout] u_bdry_contr: Contributions to the zonal ice[inout] v_bdry_contr: Contributions to the zonal ice
-
subroutine
calc_shelf_visc(CS CS, ISS ISS, G G, US US, u u, v v)¶ Update depth integrated viscosity, based on horizontal strain rates, and also update the nonlinear part of the basal traction.
- Parameters
[inout] cs: A pointer to the ice shelf control structure[in] iss: A structure with elements that describe the ice-shelf state[in] g: The grid structure used by the ice shelf.[in] us: Pointer to a structure containing unit conversion factors[inout] u: The zonal ice shelf velocity [m year-1].[inout] v: The meridional ice shelf velocity [m year-1].
-
subroutine
update_od_ffrac(CS CS, G G, US US, ocean_mass ocean_mass, find_avg find_avg)¶ - Parameters
[inout] cs: A pointer to the ice shelf control structure[inout] g: The grid structure used by the ice shelf.[in] us: Pointer to a structure containing unit conversion factors[in] ocean_mass: The mass per unit area of the ocean [kg m-2].[in] find_avg: If true, find the average of OD and ffrac, and reset the underlying running sums to 0.
-
subroutine
update_od_ffrac_uncoupled(CS CS, G G, h_shelf h_shelf)¶ - Parameters
[inout] cs: A pointer to the ice shelf control structure[in] g: The grid structure used by the ice shelf.[in] h_shelf: the thickness of the ice shelf [Z ~> m].
-
subroutine
bilinear_shape_functions(X X, Y Y, Phi Phi, area area)¶ This subroutine calculates the gradients of bilinear basis elements that that are centered at the vertices of the cell. values are calculated at points of gaussian quadrature.
- Parameters
[in] x: The x-positions of the vertices of the quadrilateral.[in] y: The y-positions of the vertices of the quadrilateral.[inout] phi: The gradients of bilinear basis elements at Gaussian quadrature points surrounding the cell verticies.[out] area: The quadrilateral cell area [m2].
-
subroutine
bilinear_shape_functions_subgrid(Phisub Phisub, nsub nsub)¶ - Parameters
[inout] phisub: Quadrature structure weights at subgridscale[in] nsub: The nubmer of subgridscale quadrature locations in each direction
-
subroutine
update_velocity_masks(CS CS, G G, hmask hmask, umask umask, vmask vmask, u_face_mask u_face_mask, v_face_mask v_face_mask)¶ - Parameters
[in] cs: A pointer to the ice shelf dynamics control structure[inout] g: The grid structure used by the ice shelf.[in] hmask: A mask indicating which tracer points are[out] umask: A coded mask indicating the nature of the[out] vmask: A coded mask indicating the nature of the[out] u_face_mask: A coded mask for velocities at the C-grid u-face[out] v_face_mask: A coded mask for velocities at the C-grid v-face
-
subroutine
interpolate_h_to_b(G G, h_shelf h_shelf, hmask hmask, H_node H_node)¶ Interpolate the ice shelf thickness from tracer point to nodal points, subject to a mask.
- Parameters
[inout] g: The grid structure used by the ice shelf.[in] h_shelf: The ice shelf thickness at tracer points [Z ~> m].[in] hmask: A mask indicating which tracer points are[inout] h_node: The ice shelf thickness at nodal (corner)
-
subroutine, public mom_ice_shelf_dynamics::ice_shelf_dyn_end(CS CS) Deallocates all memory associated with the ice shelf dynamics module.
- Parameters
cs: A pointer to the ice shelf dynamics control structure
-
subroutine
ice_shelf_temp(CS CS, ISS ISS, G G, US US, time_step time_step, melt_rate melt_rate, Time Time)¶ This subroutine updates the vertically averaged ice shelf temperature.
- Parameters
[inout] cs: A pointer to the ice shelf control structure[in] iss: A structure with elements that describe the ice-shelf state[inout] g: The grid structure used by the ice shelf.[in] us: Pointer to a structure containing unit conversion factors[in] time_step: The time step for this update [s].[in] melt_rate: basal melt rate [kg m-2 s-1][in] time: The current model time
-
subroutine
ice_shelf_advect_temp_x(CS CS, G G, time_step time_step, hmask hmask, h0 h0, h_after_uflux h_after_uflux, flux_enter flux_enter)¶ - Parameters
[in] cs: A pointer to the ice shelf control structure[inout] g: The grid structure used by the ice shelf.[in] time_step: The time step for this update [s].[in] hmask: A mask indicating which tracer points are[in] h0: The initial ice shelf thicknesses [Z ~> m].[inout] h_after_uflux: The ice shelf thicknesses after[inout] flux_enter: The integrated temperature flux into
-
subroutine
ice_shelf_advect_temp_y(CS CS, G G, time_step time_step, hmask hmask, h_after_uflux h_after_uflux, h_after_vflux h_after_vflux, flux_enter flux_enter)¶ - Parameters
[in] cs: A pointer to the ice shelf control structure[in] g: The grid structure used by the ice shelf.[in] time_step: The time step for this update [s].[in] hmask: A mask indicating which tracer points are[in] h_after_uflux: The ice shelf thicknesses after[inout] h_after_vflux: The ice shelf thicknesses after[inout] flux_enter: The integrated temperature flux into
-
-
namespace
mom_ice_shelf_initialize¶ Initialize ice shelf variables.
Functions
-
subroutine, public mom_ice_shelf_initialize::initialize_ice_thickness(h_shelf h_shelf, area_shelf_h area_shelf_h, hmask hmask, G G, US US, PF PF) Initialize ice shelf thickness.
- Parameters
[in] g: The ocean’s grid structure[inout] h_shelf: The ice shelf thickness [Z ~> m].[inout] area_shelf_h: The area per cell covered by the ice shelf [m2].[inout] hmask: A mask indicating which tracer points are[in] us: A structure containing unit conversion factors[in] pf: A structure to parse for run-time parameters
-
subroutine
initialize_ice_thickness_from_file(h_shelf h_shelf, area_shelf_h area_shelf_h, hmask hmask, G G, US US, PF PF)¶ Initialize ice shelf thickness from file.
- Parameters
[in] g: The ocean’s grid structure[inout] h_shelf: The ice shelf thickness [m].[inout] area_shelf_h: The area per cell covered by the ice shelf [m2].[inout] hmask: A mask indicating which tracer points are[in] us: A structure containing unit conversion factors[in] pf: A structure to parse for run-time parameters
-
subroutine
initialize_ice_thickness_channel(h_shelf h_shelf, area_shelf_h area_shelf_h, hmask hmask, G G, US US, PF PF)¶ Initialize ice shelf thickness for a channel configuration.
- Parameters
[in] g: The ocean’s grid structure[inout] h_shelf: The ice shelf thickness [Z ~> m].[inout] area_shelf_h: The area per cell covered by the ice shelf [m2].[inout] hmask: A mask indicating which tracer points are[in] us: A structure containing unit conversion factors[in] pf: A structure to parse for run-time parameters
-
-
namespace
mom_ice_shelf_state¶ Implements the thermodynamic aspects of ocean / ice-shelf interactions, along with a crude placeholder for a later implementation of full ice shelf dynamics, all using the MOM framework and coding style.
Functions
-
subroutine, public mom_ice_shelf_state::ice_shelf_state_init(ISS ISS, G G) Deallocates all memory associated with this module.
- Parameters
iss: A pointer to the ice shelf state structure[in] g: The grid structure used by the ice shelf.
-
subroutine, public mom_ice_shelf_state::ice_shelf_state_end(ISS ISS) Deallocates all memory associated with this module.
- Parameters
iss: A pointer to the ice shelf state structure
-
-
namespace
mom_int_tide_input¶ Calculates energy input to the internal tides.
Unnamed Group
-
subroutine, public mom_int_tide_input::set_int_tide_input(u u, v v, h h, tv tv, fluxes fluxes, itide itide, dt dt, G G, GV GV, US US, CS CS) Sets the model-state dependent internal tide energy sources.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[in] u: The zonal velocity [m s-1][in] v: The meridional velocity [m s-1][in] h: Layer thicknesses [H ~> m or kg m-2][in] tv: A structure containing pointers to the thermodynamic fields[in] fluxes: A structure of thermodynamic surface fluxes[inout] itide: A structure containing fields related to the internal tide sources.[in] dt: The time increment [s].cs: This module’s control structure.
-
subroutine
find_n2_bottom(h h, tv tv, T_f T_f, S_f S_f, h2 h2, fluxes fluxes, G G, GV GV, US US, N2_bot N2_bot)¶ Estimates the near-bottom buoyancy frequency (N^2).
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[in] h: Layer thicknesses [H ~> m or kg m-2][in] tv: A structure containing pointers to the thermodynamic fields[in] t_f: Temperature after vertical filtering to smooth out the values in thin layers [degC].[in] s_f: Salinity after vertical filtering to smooth out the values in thin layers [ppt].[in] h2: Bottom topographic roughness [Z2 ~> m2].[in] fluxes: A structure of thermodynamic surface fluxes[out] n2_bot: The squared buoyancy freqency at the ocean bottom [s-2].
-
subroutine, public mom_int_tide_input::int_tide_input_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS, itide itide) Initializes the data related to the internal tide input module.
- Parameters
[in] time: The current model time[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters[inout] diag: structure used to regulate diagnostic output.cs: This module’s control structure, which is initialized here.itide: A structure containing fields related to the internal tide sources.
-
subroutine, public mom_int_tide_input::int_tide_input_end(CS CS) Deallocates any memory related to the internal tide input module.
- Parameters
cs: This module’s control structure, which is deallocated here.
-
-
namespace
mom_interface_heights¶ Functions for calculating interface heights, including free surface height.
Functions
-
subroutine
find_eta_3d(h h, tv tv, G G, GV GV, US US, eta eta, eta_bt eta_bt, halo_size halo_size, eta_to_m eta_to_m)¶ Calculates the heights of all interfaces between layers, using the appropriate form for consistency with the calculation of the pressure gradient forces. Additionally, these height may be dilated for consistency with the corresponding time-average quantity from the barotropic calculation.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] h: Layer thicknesses [H ~> m or kg m-2][in] tv: A structure pointing to various thermodynamic variables.[out] eta: layer interface heights [Z ~> m] or 1/eta_to_m m).[in] eta_bt: optional barotropic variable that gives the “correct” free surface height (Boussinesq) or total water column mass per unit area (non-Boussinesq). This is used to dilate the layer. thicknesses when calculating interfaceheights [H ~> m or kg m-2].[in] halo_size: width of halo points on which to calculate eta.[in] eta_to_m: The conversion factor from the units of eta to m; by default this is USZ_to_m.
-
subroutine
find_eta_2d(h h, tv tv, G G, GV GV, US US, eta eta, eta_bt eta_bt, halo_size halo_size, eta_to_m eta_to_m)¶ Calculates the free surface height, using the appropriate form for consistency with the calculation of the pressure gradient forces. Additionally, the sea surface height may be adjusted for consistency with the corresponding time-average quantity from the barotropic calculation.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] h: Layer thicknesses [H ~> m or kg m-2][in] tv: A structure pointing to various thermodynamic variables.[out] eta: free surface height relative to mean sea level (z=0) often [Z ~> m].[in] eta_bt: optional barotropic variable that gives the “correct” free surface height (Boussinesq) or total water column mass per unit area (non-Boussinesq) [H ~> m or kg m-2].[in] halo_size: width of halo points on which to calculate eta.[in] eta_to_m: The conversion factor from the units of eta to m; by default this is USZ_to_m.
-
subroutine
-
namespace
mom_internal_tides¶ Subroutines that use the ray-tracing equations to propagate the internal tide energy density.
- Author
Benjamin Mater & Robert Hallberg, 2015
Unnamed Group
-
subroutine, public mom_internal_tides::propagate_int_tide(h h, tv tv, cn cn, TKE_itidal_input TKE_itidal_input, vel_btTide vel_btTide, Nb Nb, dt dt, G G, GV GV, US US, CS CS) Calls subroutines in this file that are needed to refract, propagate, and dissipate energy density of the internal tide.
- Parameters
[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] h: Layer thicknesses [H ~> m or kg m-2][in] tv: Pointer to thermodynamic variables (needed for wave structure).[in] tke_itidal_input: The energy input to the internal waves [W m-2].[in] vel_bttide: Barotropic velocity read from file [m s-1].[in] nb: Near-bottom buoyancy frequency [s-1].[in] dt: Length of time over which these fluxes will be applied [s].cs: The control structure returned by a previous call to int_tide_init.[in] cn: The internal wave speeds of each mode [m s-1].
-
subroutine
sum_en(G G, CS CS, En En, label label)¶ Checks for energy conservation on computational domain.
- Parameters
[in] g: The ocean’s grid structure.cs: The control structure returned by a previous call to int_tide_init.[in] en: The energy density of the internal tides [J m-2].[in] label: A label to use in error messages
-
subroutine
itidal_lowmode_loss(G G, US US, CS CS, Nb Nb, Ub Ub, En En, TKE_loss_fixed TKE_loss_fixed, TKE_loss TKE_loss, dt dt, full_halos full_halos)¶ Calculates the energy lost from the propagating internal tide due to scattering over small-scale roughness along the lines of Jayne & St. Laurent (2001).
- Parameters
[in] g: The ocean’s grid structure.[in] us: A dimensional unit scaling typecs: The control structure returned by a previous call to int_tide_init.[in] nb: Near-bottom stratification [s-1].[inout] ub: RMS (over one period) near-bottom horizontal[in] tke_loss_fixed: Fixed part of energy loss [kg Z-2 ~> kg m-2][inout] en: Energy density of the internal waves [J m-2].[out] tke_loss: Energy loss rate [W m-2][in] dt: Time increment [s].[in] full_halos: If true, do the calculation over the entirecomputational domain.
-
subroutine, public mom_internal_tides::get_lowmode_loss(i i, j j, G G, CS CS, mechanism mechanism, TKE_loss_sum TKE_loss_sum) This subroutine extracts the energy lost from the propagating internal which has been summed across all angles, frequencies, and modes for a given mechanism and location.
It can be called from another module to get values from this module’s (private) CS.
- Parameters
[in] i: The i-index of the value to be reported.[in] j: The j-index of the value to be reported.[in] g: The ocean’s grid structurecs: The control structure returned by a previous call to int_tide_init.[in] mechanism: The named mechanism of loss to return[out] tke_loss_sum: Total energy loss rate due to specified mechanism [W m-2].
-
subroutine
refract(En En, cn cn, freq freq, dt dt, G G, US US, NAngle NAngle, use_PPMang use_PPMang)¶ Implements refraction on the internal waves at a single frequency.
- Parameters
[in] g: The ocean’s grid structure.[in] nangle: The number of wave orientations in the discretized wave energy spectrum.[inout] en: The internal gravity wave energy density as a[in] cn: Baroclinic mode speed [m s-1].[in] freq: Wave frequency [s-1].[in] dt: Time step [s].[in] us: A dimensional unit scaling type[in] use_ppmang: If true, use PPM for advection rather than upwind.
-
subroutine
ppm_angular_advect(En2d En2d, CFL_ang CFL_ang, Flux_En Flux_En, NAngle NAngle, dt dt, halo_ang halo_ang)¶ This subroutine calculates the 1-d flux for advection in angular space using a monotonic piecewise parabolic scheme. This needs to be called from within i and j spatial loops.
- Parameters
[in] nangle: The number of wave orientations in the discretized wave energy spectrum.[in] dt: Time increment [s].[in] halo_ang: The halo size in angular space[in] en2d: The internal gravity wave energy density as a[in] cfl_ang: The CFL number of the energy advection across angles[out] flux_en: The time integrated internal wave energy flux across angles [J m-2 radian-1].
-
subroutine
propagate(En En, cn cn, freq freq, dt dt, G G, US US, CS CS, NAngle NAngle)¶ Propagates internal waves at a single frequency.
- Parameters
[inout] g: The ocean’s grid structure.[in] nangle: The number of wave orientations in the discretized wave energy spectrum.[inout] en: The internal gravity wave energy density as a[in] cn: Baroclinic mode speed [m s-1].[in] freq: Wave frequency [s-1].[in] dt: Time step [s].[in] us: A dimensional unit scaling typecs: The control structure returned by a previous call to int_tide_init.
-
subroutine
propagate_corner_spread(En En, energized_wedge energized_wedge, NAngle NAngle, speed speed, dt dt, G G, CS CS, LB LB)¶ This subroutine does first-order corner advection. It was written with the hopes of smoothing out the garden sprinkler effect, but is too numerically diffusive to be of much use as of yet. It is not yet compatible with reflection schemes (BDM).
- Parameters
[in] g: The ocean’s grid structure.[inout] en: The energy density integrated over an angular[in] speed: The magnitude of the group velocity at the cell[in] energized_wedge: Index of current ray direction.[in] nangle: The number of wave orientations in the discretized wave energy spectrum.[in] dt: Time increment [s].cs: The control structure returned by a previous call to continuity_PPM_init.[in] lb: A structure with the active energy loop bounds.
-
subroutine
propagate_x(En En, speed_x speed_x, Cgx_av Cgx_av, dCgx dCgx, dt dt, G G, Nangle Nangle, CS CS, LB LB)¶ Propagates the internal wave energy in the logical x-direction.
- Parameters
[in] g: The ocean’s grid structure.[in] nangle: The number of wave orientations in the discretized wave energy spectrum.[inout] en: The energy density integrated over an angular[in] speed_x: The magnitude of the group velocity at the[in] cgx_av: The average x-projection in each angular band.[in] dcgx: The difference in x-projections between the edges of each angular band.[in] dt: Time increment [s].cs: The control structure returned by a previous call to continuity_PPM_init.[in] lb: A structure with the active energy loop bounds.
-
subroutine
propagate_y(En En, speed_y speed_y, Cgy_av Cgy_av, dCgy dCgy, dt dt, G G, Nangle Nangle, CS CS, LB LB)¶ Propagates the internal wave energy in the logical y-direction.
- Parameters
[in] g: The ocean’s grid structure.[in] nangle: The number of wave orientations in the discretized wave energy spectrum.[inout] en: The energy density integrated over an angular[in] speed_y: The magnitude of the group velocity at the[in] cgy_av: The average y-projection in each angular band.[in] dcgy: The difference in y-projections between the edges of each angular band.[in] dt: Time increment [s].cs: The control structure returned by a previous call to continuity_PPM_init.[in] lb: A structure with the active energy loop bounds.
-
subroutine
zonal_flux_en(u u, h h, hL hL, hR hR, uh uh, dt dt, G G, j j, ish ish, ieh ieh, vol_CFL vol_CFL)¶ Evaluates the zonal mass or volume fluxes in a layer.
- Parameters
[in] g: The ocean’s grid structure.[in] u: The zonal velocity [m s-1].[in] h: Energy density used to calculate the fluxes [J m-2].[in] hl: Left- Energy densities in the reconstruction [J m-2].[in] hr: Right- Energy densities in the reconstruction [J m-2].[inout] uh: The zonal energy transport [J s-1].[in] dt: Time increment [s].[in] j: The j-index to work on.[in] ish: The start i-index range to work on.[in] ieh: The end i-index range to work on.[in] vol_cfl: If true, rescale the ratio of face areas to the cell areas when estimating the CFL number.
-
subroutine
merid_flux_en(v v, h h, hL hL, hR hR, vh vh, dt dt, G G, J J, ish ish, ieh ieh, vol_CFL vol_CFL)¶ Evaluates the meridional mass or volume fluxes in a layer.
- Parameters
[in] g: The ocean’s grid structure.[in] v: The meridional velocity [m s-1].[in] h: Energy density used to calculate the fluxes [J m-2].[in] hl: Left- Energy densities in the reconstruction [J m-2].[in] hr: Right- Energy densities in the reconstruction [J m-2].[inout] vh: The meridional energy transport [J s-1].[in] dt: Time increment [s].[in] j: The j-index to work on.[in] ish: The start i-index range to work on.[in] ieh: The end i-index range to work on.[in] vol_cfl: If true, rescale the ratio of face areas to the cell areas when estimating the CFL number.
-
subroutine
reflect(En En, NAngle NAngle, CS CS, G G, LB LB)¶ Reflection of the internal waves at a single frequency.
- Parameters
[in] g: The ocean’s grid structure[in] nangle: The number of wave orientations in the discretized wave energy spectrum.[inout] en: The internal gravity wave energy density as acs: The control structure returned by a previous call to int_tide_init.[in] lb: A structure with the active energy loop bounds.
-
subroutine
teleport(En En, NAngle NAngle, CS CS, G G, LB LB)¶ Moves energy across lines of partial reflection to prevent reflection of energy that is supposed to get across.
- Parameters
[in] g: The ocean’s grid structure.[in] nangle: The number of wave orientations in the discretized wave energy spectrum.[inout] en: The internal gravity wave energy density as acs: The control structure returned by a previous call to int_tide_init.[in] lb: A structure with the active energy loop bounds.
-
subroutine
correct_halo_rotation(En En, test test, G G, NAngle NAngle)¶ Rotates points in the halos where required to accommodate changes in grid orientation, such as at the tripolar fold.
- Parameters
[in] g: The ocean’s grid structure[inout] en: The internal gravity wave energy density as a function of space, angular orientation, frequency, and vertical mode [J m-2 radian-1].[in] test: An x-unit vector that has been passed through[in] nangle: The number of wave orientations in the discretized wave energy spectrum.
-
subroutine
ppm_reconstruction_x(h_in h_in, h_l h_l, h_r h_r, G G, LB LB, simple_2nd simple_2nd)¶ Calculates left/right edge values for PPM reconstruction in x-direction.
- Parameters
[in] g: The ocean’s grid structure.[in] h_in: Energy density in a sector (2D).[out] h_l: Left edge value of reconstruction (2D).[out] h_r: Right edge value of reconstruction (2D).[in] lb: A structure with the active loop bounds.[in] simple_2nd: If true, use the arithmetic mean energy densities as default edge values for a simple 2nd order scheme.
-
subroutine
ppm_reconstruction_y(h_in h_in, h_l h_l, h_r h_r, G G, LB LB, simple_2nd simple_2nd)¶ Calculates left/right edge valus for PPM reconstruction in y-direction.
- Parameters
[in] g: The ocean’s grid structure.[in] h_in: Energy density in a sector (2D).[out] h_l: Left edge value of reconstruction (2D).[out] h_r: Right edge value of reconstruction (2D).[in] lb: A structure with the active loop bounds.[in] simple_2nd: If true, use the arithmetic mean energy densities as default edge values for a simple 2nd order scheme.
-
subroutine
ppm_limit_pos(h_in h_in, h_L h_L, h_R h_R, h_min h_min, G G, iis iis, iie iie, jis jis, jie jie)¶ Limits the left/right edge values of the PPM reconstruction to give a reconstruction that is positive-definite. Here this is reinterpreted as giving a constant thickness if the mean thickness is less than h_min, with a minimum of h_min otherwise.
- Parameters
[in] g: The ocean’s grid structure.[in] h_in: Thickness of layer (2D).[inout] h_l: Left edge value (2D).[inout] h_r: Right edge value (2D).[in] h_min: The minimum thickness that can be obtained by a concave parabolic fit.[in] iis: Start i-index for computations[in] iie: End i-index for computations[in] jis: Start j-index for computations[in] jie: End j-index for computations
-
subroutine, public mom_internal_tides::internal_tides_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS) This subroutine initializes the internal tides module.
- Parameters
[in] time: The current model time.[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters.[in] diag: A structure that is used to regulate diagnostic output.cs: A pointer that is set to point to the control structure for this module.
-
subroutine, public mom_internal_tides::internal_tides_end(CS CS) This subroutine deallocates the memory associated with the internal tides control structure.
- Parameters
cs: A pointer to the control structure returned by a previous call to internal_tides_init, it will be deallocated here.
-
namespace
mom_intrinsic_functions¶ A module with intrinsic functions that are used by MOM but are not supported by some compilers.
Functions
-
real function, public mom_intrinsic_functions::invcosh(x x) Evaluate the inverse cosh, either using a math library or an equivalent expression.
- Parameters
[in] x: The argument of the inverse of cosh. NaNs will occur if x<1, but there is no error checking
-
-
namespace
mom_io¶ This module contains I/O framework code.
This file contains a number of subroutines that manipulate NetCDF files and handle input and output of fields. These subroutines, along with their purpose, are:
create_file: create a new file and set up structures that are needed for subsequent output and write out the coordinates.
reopen_file: reopen an existing file for writing and set up structures that are needed for subsequent output.
open_input_file: open the indicated file for reading only.
close_file: close an open file.
synch_file: flush the buffers, completing all pending output.
write_field: write a field to an open file.
write_time: write a value of the time axis to an open file.
read_data: read a variable from an open file.
read_time: read a time from an open file.
name_output_file: provide a name for an output file based on a name root and the time of the output.
find_input_file: find a file that has been previously written by MOM and named by name_output_file and open it for reading.
handle_error: write an error code and quit.
Functions
-
subroutine, public mom_io::create_file(unit unit, filename filename, vars vars, novars novars, fields fields, threading threading, timeunit timeunit, G G, dG dG, GV GV, checksums checksums) Routine creates a new NetCDF file. It also sets up structures that describe this file and variables that will later be written to this file. Type for describing a variable, typically a tracer.
- Parameters
[out] unit: unit id of an open file or -1 on a nonwriting PE with single file output[in] filename: full path to the file to create[in] vars: structures describing fields written to filename[in] novars: number of fields written to filename[inout] fields: array of fieldtypes for each variable[in] threading: SINGLE_FILE or MULTIPLE[in] timeunit: length of the units for time [s]. The default value is 86400.0, for 1 day.[in] g: ocean horizontal grid structure; G or dG is required if the new file uses any horizontal grid axes.[in] dg: dynamic horizontal grid structure; G or dG is required if the new file uses any horizontal grid axes.[in] gv: ocean vertical grid structure, which is required if the new file uses any vertical grid axes.[in] checksums: checksums of vars
-
subroutine, public mom_io::reopen_file(unit unit, filename filename, vars vars, novars novars, fields fields, threading threading, timeunit timeunit, G G, dG dG, GV GV) This routine opens an existing NetCDF file for output. If it does not find the file, a new file is created. It also sets up structures that describe this file and the variables that will later be written to this file.
- Parameters
[out] unit: unit id of an open file or -1 on a nonwriting PE with single file output[in] filename: full path to the file to create[in] vars: structures describing fields written to filename[in] novars: number of fields written to filename[inout] fields: array of fieldtypes for each variable[in] threading: SINGLE_FILE or MULTIPLE[in] timeunit: length of the units for time [s]. The default value is 86400.0, for 1 day.[in] g: ocean horizontal grid structure; G or dG is required if a new file uses any horizontal grid axes.[in] dg: dynamic horizontal grid structure; G or dG is required if a new file uses any horizontal grid axes.[in] gv: ocean vertical grid structure, which is required if a new file uses any vertical grid axes.
-
subroutine, public mom_io::read_axis_data(filename filename, axis_name axis_name, var var) Read the data associated with a named axis in a file.
- Parameters
[in] filename: Name of the file to read[in] axis_name: Name of the axis to read[out] var: The axis location data
-
integer function, public mom_io::num_timelevels(filename filename, varname varname, min_dims min_dims) This function determines how many time levels a variable has.
- Return
number of time levels varname has in filename
- Parameters
[in] filename: name of the file to read[in] varname: variable whose number of time levels are to be returned[in] min_dims: The minimum number of dimensions a variable must have if it has a time dimension. If the variable has 1 less dimension than this, then 0 is returned.
-
type(vardesc) function, public mom_io::var_desc(name name, units units, longname longname, hor_grid hor_grid, z_grid z_grid, t_grid t_grid, cmor_field_name cmor_field_name, cmor_units cmor_units, cmor_longname cmor_longname, conversion conversion, caller caller) Returns a vardesc type whose elements have been filled with the provided fields. The argument name is required, while the others are optional and have default values that are empty strings or are appropriate for a 3-d tracer field at the tracer cell centers.
- Return
vardesc type that is created
- Parameters
[in] name: variable name[in] units: variable units[in] longname: variable long name[in] hor_grid: variable horizonal staggering[in] z_grid: variable vertical staggering[in] t_grid: time description: s, p, or 1[in] cmor_field_name: CMOR name[in] cmor_units: CMOR physical dimensions of variable[in] cmor_longname: CMOR long name[in] conversion: for unit conversions, such as needed to convert from intensive to extensive[in] caller: calling routine?
-
subroutine, public mom_io::modify_vardesc(vd vd, name name, units units, longname longname, hor_grid hor_grid, z_grid z_grid, t_grid t_grid, cmor_field_name cmor_field_name, cmor_units cmor_units, cmor_longname cmor_longname, conversion conversion, caller caller) This routine modifies the named elements of a vardesc type. All arguments are optional, except the vardesc type to be modified.
- Parameters
[inout] vd: vardesc type that is modified[in] name: name of variable[in] units: units of variable[in] longname: long name of variable[in] hor_grid: horizonal staggering of variable[in] z_grid: vertical staggering of variable[in] t_grid: time description: s, p, or 1[in] cmor_field_name: CMOR name[in] cmor_units: CMOR physical dimensions of variable[in] cmor_longname: CMOR long name[in] conversion: for unit conversions, such as needed to convert from intensive to extensive[in] caller: calling routine?
-
character(len=len(longname)) function, public mom_io::cmor_long_std(longname longname) This function returns the CMOR standard name given a CMOR longname, based on the standard pattern of character conversions.
- Return
The CMOR standard name generated from longname
- Parameters
[in] longname: The CMOR longname being converted
-
subroutine, public mom_io::query_vardesc(vd vd, name name, units units, longname longname, hor_grid hor_grid, z_grid z_grid, t_grid t_grid, cmor_field_name cmor_field_name, cmor_units cmor_units, cmor_longname cmor_longname, conversion conversion, caller caller) This routine queries vardesc.
- Parameters
[in] vd: vardesc type that is queried[out] name: name of variable[out] units: units of variable[out] longname: long name of variable[out] hor_grid: horiz staggering of variable[out] z_grid: vert staggering of variable[out] t_grid: time description: s, p, or 1[out] cmor_field_name: CMOR name[out] cmor_units: CMOR physical dimensions of variable[out] cmor_longname: CMOR long name[out] conversion: for unit conversions, such as needed to convert from intensive to extensive[in] caller: calling routine?
-
subroutine
safe_string_copy(str1 str1, str2 str2, fieldnm fieldnm, caller caller)¶ Copies a string.
- Parameters
[in] str1: The string being copied[out] str2: The string being copied into[in] fieldnm: The name of the field for error messages[in] caller: The calling routine for error messages
-
character(len=len(name)) function, public mom_io::ensembler(name name, ens_no_in ens_no_in) Returns a name with “%#E” or “%E” replaced with the ensemble member number.
- Return
The name encoded with the ensemble number
- Parameters
[in] name: The name to be modified[in] ens_no_in: The number of the current ensemble member
-
logical function mom_io::mom_file_exists(filename filename, MOM_Domain MOM_Domain) Returns true if the named file or its domain-decomposed variant exists.
- Parameters
[in] filename: The name of the file being inquired about[in] mom_domain: The MOM_Domain that describes the decomposition
-
logical function mom_io::fms_file_exists(filename filename, domain domain, no_domain no_domain) Returns true if the named file or its domain-decomposed variant exists.
- Parameters
[in] filename: The name of the file being inquired about[in] domain: The mpp domain2d that describes the decomposition[in] no_domain: This file does not use domain decomposition
-
subroutine
mom_read_data_1d(filename filename, fieldname fieldname, data data, timelevel timelevel, scale scale)¶ This function uses the fms_io function read_data to read 1-D data field named “fieldname” from file “filename”.
- Parameters
[in] filename: The name of the file to read[in] fieldname: The variable name of the data in the file[inout] data: The 1-dimensional array into which the data[in] timelevel: The time level in the file to read[in] scale: A scaling factor that the field is multiplied by before they are returned.
-
subroutine
mom_read_data_2d(filename filename, fieldname fieldname, data data, MOM_Domain MOM_Domain, timelevel timelevel, position position, scale scale)¶ This function uses the fms_io function read_data to read a distributed 2-D data field named “fieldname” from file “filename”. Valid values for “position” include CORNER, CENTER, EAST_FACE and NORTH_FACE.
- Parameters
[in] filename: The name of the file to read[in] fieldname: The variable name of the data in the file[inout] data: The 2-dimensional array into which the data should be read[in] mom_domain: The MOM_Domain that describes the decomposition[in] timelevel: The time level in the file to read[in] position: A flag indicating where this data is located[in] scale: A scaling factor that the field is multiplied by before they are returned.
-
subroutine
mom_read_data_3d(filename filename, fieldname fieldname, data data, MOM_Domain MOM_Domain, timelevel timelevel, position position, scale scale)¶ This function uses the fms_io function read_data to read a distributed 3-D data field named “fieldname” from file “filename”. Valid values for “position” include CORNER, CENTER, EAST_FACE and NORTH_FACE.
- Parameters
[in] filename: The name of the file to read[in] fieldname: The variable name of the data in the file[inout] data: The 3-dimensional array into which the data should be read[in] mom_domain: The MOM_Domain that describes the decomposition[in] timelevel: The time level in the file to read[in] position: A flag indicating where this data is located[in] scale: A scaling factor that the field is multiplied by before they are returned.
-
subroutine
mom_read_data_4d(filename filename, fieldname fieldname, data data, MOM_Domain MOM_Domain, timelevel timelevel, position position, scale scale)¶ This function uses the fms_io function read_data to read a distributed 4-D data field named “fieldname” from file “filename”. Valid values for “position” include CORNER, CENTER, EAST_FACE and NORTH_FACE.
- Parameters
[in] filename: The name of the file to read[in] fieldname: The variable name of the data in the file[inout] data: The 4-dimensional array into which the data should be read[in] mom_domain: The MOM_Domain that describes the decomposition[in] timelevel: The time level in the file to read[in] position: A flag indicating where this data is located[in] scale: A scaling factor that the field is multiplied by before they are returned.
-
subroutine
mom_read_vector_2d(filename filename, u_fieldname u_fieldname, v_fieldname v_fieldname, u_data u_data, v_data v_data, MOM_Domain MOM_Domain, timelevel timelevel, stagger stagger, scalar_pair scalar_pair, scale scale)¶ This function uses the fms_io function read_data to read a pair of distributed 2-D data fields with names given by “[uv]_fieldname” from file “filename”. Valid values for “stagger” include CGRID_NE, BGRID_NE, and AGRID.
- Parameters
[in] filename: The name of the file to read[in] u_fieldname: The variable name of the u data in the file[in] v_fieldname: The variable name of the v data in the file[inout] u_data: The 2 dimensional array into which the u-component of the data should be read[inout] v_data: The 2 dimensional array into which the v-component of the data should be read[in] mom_domain: The MOM_Domain that describes the decomposition[in] timelevel: The time level in the file to read[in] stagger: A flag indicating where this vector is discretized[in] scalar_pair: If true, a pair of scalars are to be read.cretized[in] scale: A scaling factor that the fields are multiplied by before they are returned.
-
subroutine
mom_read_vector_3d(filename filename, u_fieldname u_fieldname, v_fieldname v_fieldname, u_data u_data, v_data v_data, MOM_Domain MOM_Domain, timelevel timelevel, stagger stagger, scalar_pair scalar_pair, scale scale)¶ This function uses the fms_io function read_data to read a pair of distributed 3-D data fields with names given by “[uv]_fieldname” from file “filename”. Valid values for “stagger” include CGRID_NE, BGRID_NE, and AGRID.
- Parameters
[in] filename: The name of the file to read[in] u_fieldname: The variable name of the u data in the file[in] v_fieldname: The variable name of the v data in the file[inout] u_data: The 3 dimensional array into which the u-component of the data should be read[inout] v_data: The 3 dimensional array into which the v-component of the data should be read[in] mom_domain: The MOM_Domain that describes the decomposition[in] timelevel: The time level in the file to read[in] stagger: A flag indicating where this vector is discretized[in] scalar_pair: If true, a pair of scalars are to be read.cretized[in] scale: A scaling factor that the fields are multiplied by before they are returned.
-
subroutine, public mom_io::mom_io_init(param_file param_file) Initialize the MOM_io module.
- Parameters
[in] param_file: structure indicating the open file to parse for model parameter values.
-
namespace
mom_isopycnal_slopes¶ Calculations of isoneutral slopes and stratification.
Functions
-
subroutine, public mom_isopycnal_slopes::calc_isoneutral_slopes(G G, GV GV, US US, h h, e e, tv tv, dt_kappa_smooth dt_kappa_smooth, slope_x slope_x, slope_y slope_y, N2_u N2_u, N2_v N2_v, halo halo) Calculate isopycnal slopes, and optionally return N2 used in calculation.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[in] h: Layer thicknesses [H ~> m or kg m-2][in] e: Interface heights [Z ~> m] or units given by 1/eta_to_m)[in] tv: A structure pointing to various thermodynamic variables[in] dt_kappa_smooth: A smoothing vertical diffusivity times a smoothing timescale [Z2 ~> m2].[inout] slope_x: Isopycnal slope in i-direction [nondim][inout] slope_y: Isopycnal slope in j-direction [nondim][inout] n2_u: Brunt-Vaisala frequency squared at[inout] n2_v: Brunt-Vaisala frequency squared at[in] halo: Halo width over which to compute
-
subroutine, public mom_isopycnal_slopes::vert_fill_ts(h h, T_in T_in, S_in S_in, kappa_dt kappa_dt, T_f T_f, S_f S_f, G G, GV GV, halo_here halo_here, larger_h_denom larger_h_denom) Returns tracer arrays (nominally T and S) with massless layers filled with sensible values, by diffusing vertically with a small but constant diffusivity.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][in] t_in: Input temperature [degC][in] s_in: Input salinity [ppt][in] kappa_dt: A vertical diffusivity to use for smoothing times a smoothing timescale [Z2 ~> m2].[out] t_f: Filled temperature [degC][out] s_f: Filled salinity [ppt][in] halo_here: Number of halo points to work on, 0 by default[in] larger_h_denom: Present and true, add a large enough minimal thickness in the denominator of the flux calculations so that the fluxes are never so large as eliminate the transmission of information across groups of massless layers.
-
-
namespace
mom_kappa_shear¶ Shear-dependent mixing following Jackson et al. 2008.
By Laura Jackson and Robert Hallberg, 2006-2008
This file contains the subroutines that determine the diapycnal diffusivity driven by resolved shears, as specified by the parameterizations described in Jackson and Hallberg (JPO, 2008).
The technique by which the 6 equations (for kappa, TKE, u, v, T, and S) are solved simultaneously has been dramatically revised from the previous version. The previous version was not converging in some cases, especially near the surface mixed layer, while the revised version does. The revised version solves for kappa and TKE with shear and stratification fixed, then marches the density and velocities forward with an adaptive (and aggressive) time step in a predictor-corrector-corrector emulation of a trapezoidal scheme. Run-time-settable parameters determine the tolerence to which the kappa and TKE equations are solved and the minimum time step that can be taken.
Unnamed Group
-
subroutine, public mom_kappa_shear::calculate_kappa_shear(u_in u_in, v_in v_in, h h, tv tv, p_surf p_surf, kappa_io kappa_io, tke_io tke_io, kv_io kv_io, dt dt, G G, GV GV, US US, CS CS, initialize_all initialize_all) Subroutine for calculating shear-driven diffusivity and TKE in tracer columns.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] u_in: Initial zonal velocity [m s-1]. (Intent in)[in] v_in: Initial meridional velocity [m s-1].[in] h: Layer thicknesses [H ~> m or kg m-2].[in] tv: A structure containing pointers to any available thermodynamic fields. Absent fields have NULL ptrs.p_surf: The pressure at the ocean surface [Pa] (or NULL).[inout] kappa_io: The diapycnal diffusivity at each interface[out] tke_io: The turbulent kinetic energy per unit mass at[inout] kv_io: The vertical viscosity at each interface[in] dt: Time increment [T ~> s].cs: The control structure returned by a previous call to kappa_shear_init.[in] initialize_all: If present and false, the previous value of kappa is used to start the iterations
-
subroutine, public mom_kappa_shear::calc_kappa_shear_vertex(u_in u_in, v_in v_in, h h, T_in T_in, S_in S_in, tv tv, p_surf p_surf, kappa_io kappa_io, tke_io tke_io, kv_io kv_io, dt dt, G G, GV GV, US US, CS CS, initialize_all initialize_all) Subroutine for calculating shear-driven diffusivity and TKE in corner columns.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] u_in: Initial zonal velocity [m s-1]. (Intent in)[in] v_in: Initial meridional velocity [m s-1].[in] h: Layer thicknesses [H ~> m or kg m-2].[in] t_in: Layer potential temperatures [degC][in] s_in: Layer salinities in ppt.[in] tv: A structure containing pointers to any available thermodynamic fields. Absent fields have NULL ptrs.p_surf: The pressure at the ocean surface [Pa] (or NULL).[out] kappa_io: The diapycnal diffusivity at each interface[out] tke_io: The turbulent kinetic energy per unit mass at[inout] kv_io: The vertical viscosity at each interface [Z2 T-1 ~> m2 s-1].[in] dt: Time increment [T ~> s].cs: The control structure returned by a previous call to kappa_shear_init.[in] initialize_all: If present and false, the previous value of kappa is used to start the iterations
-
subroutine
kappa_shear_column(kappa kappa, tke tke, dt dt, nzc nzc, f2 f2, surface_pres surface_pres, dz dz, u0xdz u0xdz, v0xdz v0xdz, T0xdz T0xdz, S0xdz S0xdz, kappa_avg kappa_avg, tke_avg tke_avg, tv tv, CS CS, GV GV, US US, I_Ld2_1d I_Ld2_1d, dz_Int_1d dz_Int_1d)¶ This subroutine calculates shear-driven diffusivity and TKE in a single column.
- Parameters
[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[inout] kappa: The time-weighted average of kappa [Z2 T-1 ~> m2 s-1].[out] tke: The Turbulent Kinetic Energy per unit mass at[in] nzc: The number of active layers in the column.[in] f2: The square of the Coriolis parameter [T-2 ~> s-2].[in] surface_pres: The surface pressure [Pa].[in] dz: The layer thickness [Z ~> m].[in] u0xdz: The initial zonal velocity times dz [Z L T-1 ~> m2 s-1].[in] v0xdz: The initial meridional velocity times dz [Z L T-1 ~> m2 s-1].[in] t0xdz: The initial temperature times dz [degC Z ~> degC m].[in] s0xdz: The initial salinity times dz [ppt Z ~> ppt m].[out] kappa_avg: The time-weighted average of kappa [Z2 T-1 ~> m2 s-1].[out] tke_avg: The time-weighted average of TKE [Z2 T-2 ~> m2 s-2].[in] dt: Time increment [T ~> s].[in] tv: A structure containing pointers to any available thermodynamic fields. Absent fields have NULL ptrs.cs: The control structure returned by a previous call to kappa_shear_init.[out] i_ld2_1d: The inverse of the squared mixing length [Z-2 ~> m-2].[out] dz_int_1d: The extent of a finite-volume space surrounding an interface,
-
subroutine
calculate_projected_state(kappa kappa, u0 u0, v0 v0, T0 T0, S0 S0, dt dt, nz nz, dz dz, I_dz_int I_dz_int, dbuoy_dT dbuoy_dT, dbuoy_dS dbuoy_dS, u u, v v, T T, Sal Sal, GV GV, US US, N2 N2, S2 S2, ks_int ks_int, ke_int ke_int, vel_underflow vel_underflow)¶ This subroutine calculates the velocities, temperature and salinity that the water column will have after mixing for dt with diffusivities kappa. It may also calculate the projected buoyancy frequency and shear.
- Parameters
[in] nz: The number of layers (after eliminating massless layers?).[in] kappa: The diapycnal diffusivity at interfaces, [Z2 T-1 ~> m2 s-1].[in] u0: The initial zonal velocity [m s-1].[in] v0: The initial meridional velocity [m s-1].[in] t0: The initial temperature [degC].[in] s0: The initial salinity [ppt].[in] dz: The grid spacing of layers [Z ~> m].[in] i_dz_int: The inverse of the layer’s thicknesses [Z-1 ~> m-1].[in] dbuoy_dt: The partial derivative of buoyancy with temperature [Z T-2 degC-1 ~> m s-2 degC-1].[in] dbuoy_ds: The partial derivative of buoyancy with salinity [Z T-2 ppt-1 ~> m s-2 ppt-1].[in] dt: The time step [T ~> s].[inout] u: The zonal velocity after dt [m s-1].[inout] v: The meridional velocity after dt [m s-1].[inout] t: The temperature after dt [degC].[inout] sal: The salinity after dt [ppt].[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[inout] n2: The buoyancy frequency squared at interfaces [T-2 ~> s-2].[inout] s2: The squared shear at interfaces [T-2 ~> s-2].[in] ks_int: The topmost k-index with a non-zero diffusivity.[in] ke_int: The bottommost k-index with a non-zero diffusivity.[in] vel_underflow: If present and true, any velocities that are smaller in magnitude than this value are set to 0 [m s-1].
-
subroutine
find_kappa_tke(N2 N2, S2 S2, kappa_in kappa_in, Idz Idz, dz_Int dz_Int, I_L2_bdry I_L2_bdry, f2 f2, nz nz, CS CS, GV GV, US US, K_Q K_Q, tke tke, kappa kappa, kappa_src kappa_src, local_src local_src)¶ This subroutine calculates new, consistent estimates of TKE and kappa.
- Parameters
[in] nz: The number of layers to work on.[in] n2: The buoyancy frequency squared at interfaces [T-2 ~> s-2].[in] s2: The squared shear at interfaces [T-2 ~> s-2].[in] kappa_in: The initial guess at the diffusivity [Z2 T-1 ~> m2 s-1].[in] dz_int: The thicknesses associated with interfaces [Z-1 ~> m-1].[in] i_l2_bdry: The inverse of the squared distance to boundaries [Z-2 !> m-2].[in] idz: The inverse grid spacing of layers [Z-1 ~> m-1].[in] f2: The squared Coriolis parameter [T-2 ~> s-2].cs: A pointer to this module’s control structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[inout] k_q: The shear-driven diapycnal diffusivity divided by the turbulent kinetic energy per unit mass at interfaces [Z2 m-2 s2 T-1 ~> s].[out] tke: The turbulent kinetic energy per unit mass at interfaces [Z2 T-2 ~> m2 s-2].[out] kappa: The diapycnal diffusivity at interfaces [Z2 T-1 ~> m2 s-1].[out] kappa_src: The source term for kappa [T-1].[out] local_src: The sum of all local sources for kappa,
-
logical function, public mom_kappa_shear::kappa_shear_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS) This subroutineinitializesthe parameters that regulate shear-driven mixing.
- Return
True if module is to be used, False otherwise
- Parameters
[in] time: The current model time.[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters.[inout] diag: A structure that is used to regulate diagnostic output.cs: A pointer that is set to point to the control structure for this module
-
logical function, public mom_kappa_shear::kappa_shear_is_used(param_file param_file) This function indicates to other modules whether the Jackson et al shear mixing parameterization will be used without needing to duplicate the log entry.
- Parameters
[in] param_file: A structure to parse for run-time parameters
-
logical function, public mom_kappa_shear::kappa_shear_at_vertex(param_file param_file) This function indicates to other modules whether the Jackson et al shear mixing parameterization will be used without needing to duplicate the log entry.
- Parameters
[in] param_file: A structure to parse for run-time parameters
-
-
namespace
mom_lateral_mixing_coeffs¶ Variable mixing coefficients.
This module provides a container for various factors used in prescribing diffusivities, that are a function of the state (in particular the stratification and isoneutral slopes).
Functions
-
subroutine, public mom_lateral_mixing_coeffs::calc_resoln_function(h h, tv tv, G G, GV GV, US US, CS CS) Calculates and stores the non-dimensional resolution functions.
- Parameters
[inout] g: Ocean grid structure[in] h: Layer thickness [H ~> m or kg m-2][in] tv: Thermodynamic variables[in] gv: Vertical grid structure[in] us: A dimensional unit scaling typecs: Variable mixing coefficients
-
subroutine, public mom_lateral_mixing_coeffs::calc_slope_functions(h h, tv tv, dt dt, G G, GV GV, US US, CS CS) Calculates and stores functions of isopycnal slopes, e.g. Sx, Sy, S*N, mostly used in the Visbeck et al. style scaling of diffusivity.
- Parameters
[inout] g: Ocean grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[inout] h: Layer thickness [H ~> m or kg m-2][in] tv: Thermodynamic variables[in] dt: Time increment [s]cs: Variable mixing coefficients
-
subroutine
calc_visbeck_coeffs(h h, slope_x slope_x, slope_y slope_y, N2_u N2_u, N2_v N2_v, G G, GV GV, CS CS)¶ Calculates factors used when setting diffusivity coefficients similar to Visbeck et al.
- Parameters
[inout] g: Ocean grid structure[in] gv: Vertical grid structure[in] h: Layer thickness [H ~> m or kg m-2][in] slope_x: Zonal isoneutral slope[in] n2_u: Brunt-Vaisala frequency at u-points [s-2][in] slope_y: Meridional isoneutral slope[in] n2_v: Brunt-Vaisala frequency at v-points [s-2]cs: Variable mixing coefficients
-
subroutine
calc_slope_functions_using_just_e(h h, G G, GV GV, US US, CS CS, e e, calculate_slopes calculate_slopes)¶ The original calc_slope_function() that calculated slopes using interface positions only, not accounting for density variations.
- Parameters
[inout] g: Ocean grid structure[inout] h: Layer thickness [H ~> m or kg m-2][in] gv: Vertical grid structure[in] us: A dimensional unit scaling typecs: Variable mixing coefficients[in] e: Interface position [Z ~> m][in] calculate_slopes: If true, calculate slopes internally otherwise use slopes stored in CS
-
subroutine, public mom_lateral_mixing_coeffs::calc_qg_leith_viscosity(CS CS, G G, GV GV, h h, k k, div_xx_dx div_xx_dx, div_xx_dy div_xx_dy, vort_xy_dx vort_xy_dx, vort_xy_dy vort_xy_dy) Calculates the Leith Laplacian and bi-harmonic viscosity coefficients.
- Parameters
cs: Variable mixing coefficients[in] g: Ocean grid structure[in] gv: The ocean’s vertical grid structure.[inout] h: Layer thickness [H ~> m or kg m-2][in] k: Layer for which to calculate vorticity magnitude[in] div_xx_dx: x-derivative of horizontal divergence (d/dx(du/dx + dv/dy)) [m-1 s-1][in] div_xx_dy: y-derivative of horizontal divergence (d/dy(du/dx + dv/dy)) [m-1 s-1][inout] vort_xy_dx: x-derivative of vertical vorticity (d/dx(dv/dx - du/dy)) [m-1 s-1][inout] vort_xy_dy: y-derivative of vertical vorticity (d/dy(dv/dx - du/dy)) [m-1 s-1]
-
subroutine, public mom_lateral_mixing_coeffs::varmix_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS) Initializes the variables mixing coefficients container.
- Parameters
[in] time: Current model time[in] g: Ocean grid structure[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: Parameter file handles[inout] diag: Diagnostics control structurecs: Variable mixing coefficients
-
-
namespace
mom_marine_ice¶ Routines incorporating the effects of marine ice (sea-ice and icebergs) into the ocean model dynamics and thermodynamics.
Functions
-
subroutine, public mom_marine_ice::iceberg_forces(G G, forces forces, use_ice_shelf use_ice_shelf, sfc_state sfc_state, time_step time_step, CS CS) add_berg_flux_to_shelf adds rigidity and ice-area coverage due to icebergs to the forces type fields, and adds ice-areal coverage and modifies various thermodynamic fluxes due to the presence of icebergs.
- Parameters
[inout] g: The ocean’s grid structure[inout] forces: A structure with the driving mechanical forces[inout] sfc_state: A structure containing fields that describe the surface state of the ocean.[in] use_ice_shelf: If true, this configuration uses ice shelves.[in] time_step: The coupling time step [s].cs: Pointer to the control structure for MOM_marine_ice
-
subroutine, public mom_marine_ice::iceberg_fluxes(G G, fluxes fluxes, use_ice_shelf use_ice_shelf, sfc_state sfc_state, time_step time_step, CS CS) iceberg_fluxes adds ice-area-coverage and modifies various thermodynamic fluxes due to the presence of icebergs.
- Parameters
[inout] g: The ocean’s grid structure[inout] fluxes: A structure with pointers to themodynamic, tracer and mass exchange forcing fields[inout] sfc_state: A structure containing fields that describe the surface state of the ocean.[in] use_ice_shelf: If true, this configuration uses ice shelves.[in] time_step: The coupling time step [s].cs: Pointer to the control structure for MOM_marine_ice
-
subroutine, public mom_marine_ice::marine_ice_init(Time Time, G G, param_file param_file, diag diag, CS CS) Initialize control structure for MOM_marine_ice.
- Parameters
[in] time: Current model time[in] g: Ocean grid structure[in] param_file: Runtime parameter handles[inout] diag: Diagnostics control structurecs: Pointer to the control structure for MOM_marine_ice
-
-
namespace
mom_meke¶ Implements the Mesoscale Eddy Kinetic Energy framework with topographic beta effect included in computing beta in Rhines scale.
Unnamed Group
-
subroutine, public mom_meke::step_forward_meke(MEKE MEKE, h h, SN_u SN_u, SN_v SN_v, visc visc, dt dt, G G, GV GV, US US, CS CS, hu hu, hv hv) Integrates forward-in-time the MEKE eddy energy equation. See MEKE equations.
- Parameters
meke: MEKE data.[inout] g: Ocean grid.[in] gv: Ocean vertical grid structure.[in] us: A dimensional unit scaling type[in] h: Layer thickness [H ~> m or kg m-2].[inout] sn_u: Eady growth rate at u-points [s-1].[inout] sn_v: Eady growth rate at v-points [s-1].[in] visc: The vertical viscosity type.[in] dt: Model(baroclinic) time-step [s].cs: MEKE control structure.[in] hu: Zonal mass flux [H m2 s-1 ~> m3 s-1 or kg s-1].[in] hv: Meridional mass flux [H m2 s-1 ~> m3 s-1 or kg s-1]
-
subroutine
meke_equilibrium(CS CS, MEKE MEKE, G G, GV GV, US US, SN_u SN_u, SN_v SN_v, drag_rate_visc drag_rate_visc, I_mass I_mass)¶ Calculates the equilibrium solutino where the source depends only on MEKE diffusivity and there is no lateral diffusion of MEKE. Results is in MEKEMEKE.
- Parameters
[inout] g: Ocean grid.[in] gv: Ocean vertical grid structure.[in] us: A dimensional unit scaling typecs: MEKE control structure.meke: MEKE data.[in] sn_u: Eady growth rate at u-points [s-1].[in] sn_v: Eady growth rate at v-points [s-1].[in] drag_rate_visc: Mean flow contrib. to drag rate[in] i_mass: Inverse of column mass.
-
subroutine
meke_lengthscales(CS CS, MEKE MEKE, G G, GV GV, US US, SN_u SN_u, SN_v SN_v, EKE EKE, bottomFac2 bottomFac2, barotrFac2 barotrFac2, LmixScale LmixScale)¶ Calculates the eddy mixing length scale and \(\gamma_b\) and \(\gamma_t\) functions that are ratios of either bottom or barotropic eddy energy to the column eddy energy, respectively. See MEKE equations.
- Parameters
cs: MEKE control structure.meke: MEKE data.[inout] g: Ocean grid.[in] gv: Ocean vertical grid structure.[in] us: A dimensional unit scaling type[in] sn_u: Eady growth rate at u-points [s-1].[in] sn_v: Eady growth rate at v-points [s-1].[in] eke: Eddy kinetic energy [m2 s-2].[out] bottomfac2: gamma_b^2[out] barotrfac2: gamma_t^2[out] lmixscale: Eddy mixing length [m].
-
subroutine
meke_lengthscales_0d(CS CS, area area, beta beta, depth depth, Rd_dx Rd_dx, SN SN, EKE EKE, Z_to_L Z_to_L, bottomFac2 bottomFac2, barotrFac2 barotrFac2, LmixScale LmixScale, Lrhines Lrhines, Leady Leady)¶ Calculates the eddy mixing length scale and \(\gamma_b\) and \(\gamma_t\) functions that are ratios of either bottom or barotropic eddy energy to the column eddy energy, respectively. See MEKE equations.
- Parameters
cs: MEKE control structure.[in] area: Grid cell area [m2][in] beta: Planetary beta = |grad F| [s-1 m-1][in] depth: Ocean depth [Z ~> m][in] rd_dx: Resolution Ld/dx [nondim].[in] sn: Eady growth rate [s-1].[in] eke: Eddy kinetic energy [m s-1].[in] z_to_l: A conversion factor from depth units (Z) to the units for lateral distances (L).[out] bottomfac2: gamma_b^2[out] barotrfac2: gamma_t^2[out] lmixscale: Eddy mixing length [m].[out] lrhines: Rhines length scale [m].[out] leady: Eady length scale [m].
-
logical function, public mom_meke::meke_init(Time Time, G G, US US, param_file param_file, diag diag, CS CS, MEKE MEKE, restart_CS restart_CS) Initializes the MOM_MEKE module and reads parameters. Returns True if module is to be used, otherwise returns False.
- Parameters
[in] time: The current model time.[inout] g: The ocean’s grid structure.[in] us: A dimensional unit scaling type[in] param_file: Parameter file parser structure.[inout] diag: Diagnostics structure.cs: MEKE control structure.meke: MEKE-related fields.restart_cs: Restart control structure for MOM_MEKE.
-
subroutine, public mom_meke::meke_alloc_register_restart(HI HI, param_file param_file, MEKE MEKE, restart_CS restart_CS) Allocates memory and register restart fields for the MOM_MEKE module.
- Parameters
[in] hi: Horizontal index structure[in] param_file: Parameter file parser structure.meke: A structure with MEKE-related fields.restart_cs: Restart control structure for MOM_MEKE.
-
subroutine, public mom_meke::meke_end(MEKE MEKE, CS CS) Deallocates any variables allocated in MEKE_init or MEKE_alloc_register_restart.
- Parameters
meke: A structure with MEKE-related fields.cs: The control structure for MOM_MEKE.
-
-
namespace
mom_meke_types¶
-
namespace
mom_mixed_layer_restrat¶ Parameterization of mixed layer restratification by unresolved mixed-layer eddies.
Functions
-
subroutine, public mom_mixed_layer_restrat::mixedlayer_restrat(h h, uhtr uhtr, vhtr vhtr, tv tv, forces forces, dt dt, MLD MLD, VarMix VarMix, G G, GV GV, US US, CS CS) Driver for the mixed-layer restratification parameterization. The code branches between two different implementations depending on whether the bulk-mixed layer or a general coordinate are in use.
- Parameters
[inout] g: Ocean grid structure[in] gv: Ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] h: Layer thickness [H ~> m or kg m-2][inout] uhtr: Accumulated zonal mass flux [H m2 ~> m3 or kg][inout] vhtr: Accumulated meridional mass flux [H m2 ~> m3 or kg][in] tv: Thermodynamic variables structure[in] forces: A structure with the driving mechanical forces[in] dt: Time increment [s]mld: Mixed layer depth provided by the PBL scheme [H ~> m or kg m-2]varmix: Container for derived fieldscs: Module control structure
-
subroutine
mixedlayer_restrat_general(h h, uhtr uhtr, vhtr vhtr, tv tv, forces forces, dt dt, MLD_in MLD_in, VarMix VarMix, G G, GV GV, US US, CS CS)¶ Calculates a restratifying flow in the mixed layer.
- Parameters
[inout] g: Ocean grid structure[in] gv: Ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] h: Layer thickness [H ~> m or kg m-2][inout] uhtr: Accumulated zonal mass flux [H m2 ~> m3 or kg][inout] vhtr: Accumulated meridional mass flux [H m2 ~> m3 or kg][in] tv: Thermodynamic variables structure[in] forces: A structure with the driving mechanical forces[in] dt: Time increment [s]mld_in: Mixed layer depth provided by the PBL scheme [m] (not H)varmix: Container for derived fieldscs: Module control structure
-
subroutine
mixedlayer_restrat_bml(h h, uhtr uhtr, vhtr vhtr, tv tv, forces forces, dt dt, G G, GV GV, US US, CS CS)¶ Calculates a restratifying flow assuming a 2-layer bulk mixed layer.
- Parameters
[in] g: Ocean grid structure[in] gv: Ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] h: Layer thickness [H ~> m or kg m-2][inout] uhtr: Accumulated zonal mass flux [H m2 ~> m3 or kg][inout] vhtr: Accumulated meridional mass flux [H m2 ~> m3 or kg][in] tv: Thermodynamic variables structure[in] forces: A structure with the driving mechanical forces[in] dt: Time increment [s]cs: Module control structure
-
logical function, public mom_mixed_layer_restrat::mixedlayer_restrat_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS, restart_CS restart_CS) Initialize the mixed layer restratification module.
- Parameters
[in] time: Current model time[inout] g: Ocean grid structure[in] gv: Ocean vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: Parameter file to parse[inout] diag: Regulate diagnosticscs: Module control structurerestart_cs: A pointer to the restart control structure
-
subroutine, public mom_mixed_layer_restrat::mixedlayer_restrat_register_restarts(HI HI, param_file param_file, CS CS, restart_CS restart_CS) Allocate and register fields in the mixed layer restratification structure for restarts.
- Parameters
[in] hi: Horizontal index structure[in] param_file: Parameter file to parsecs: Module control structurerestart_cs: A pointer to the restart control structure
Variables
-
character(len=40) mom_mixed_layer_restrat::mdl = "MOM_mixed_layer_restrat" This module’s name.
-
-
namespace
mom_neutral_diffusion¶ A column-wise toolbox for implementing neutral diffusion.
Functions
-
logical function, public mom_neutral_diffusion::neutral_diffusion_init(Time Time, G G, param_file param_file, diag diag, EOS EOS, CS CS) Read parameters and allocate control structure for neutral_diffusion module.
- Parameters
[in] time: Time structure[in] g: Grid structure[inout] diag: Diagnostics control structure[in] param_file: Parameter file structure[in] eos: Equation of statecs: Neutral diffusion control structure
-
subroutine, public mom_neutral_diffusion::neutral_diffusion_calc_coeffs(G G, GV GV, h h, T T, S S, CS CS) Calculate remapping factors for u/v columns used to map adjoining columns to a shared coordinate space.
- Parameters
[in] g: Ocean grid structure[in] gv: ocean vertical grid structure[in] h: Layer thickness [H ~> m or kg m-2][in] t: Potential temperature [degC][in] s: Salinity [ppt]cs: Neutral diffusion control structure
-
subroutine, public mom_neutral_diffusion::neutral_diffusion(G G, GV GV, h h, Coef_x Coef_x, Coef_y Coef_y, dt dt, Reg Reg, CS CS) Update tracer concentration due to neutral diffusion; layer thickness unchanged by this update.
- Parameters
[in] g: Ocean grid structure[in] gv: ocean vertical grid structure[in] h: Layer thickness [H ~> m or kg m-2][in] coef_x: dt * Kh * dy / dx at u-points [m2][in] coef_y: dt * Kh * dx / dy at v-points [m2][in] dt: Tracer time step * I_numitts (I_numitts in tracer_hordiff)reg: Tracer registrycs: Neutral diffusion control structure
-
subroutine
interface_scalar(nk nk, h h, S S, Si Si, i_method i_method, h_neglect h_neglect)¶ Returns interface scalar, Si, for a column of layer values, S.
- Parameters
[in] nk: Number of levels[in] h: Layer thickness [H ~> m or kg m-2][in] s: Layer scalar (conc, e.g. ppt)[inout] si: Interface scalar (conc, e.g. ppt)[in] i_method: =1 use average of PLM edges =2 use continuous PPM edge interpolation[in] h_neglect: A negligibly small thickness [H ~> m or kg m-2]
-
real function mom_neutral_diffusion::ppm_edge(hkm1 hkm1, hk hk, hkp1 hkp1, hkp2 hkp2, Ak Ak, Akp1 Akp1, Pk Pk, Pkp1 Pkp1, h_neglect h_neglect) Returns the PPM quasi-fourth order edge value at k+1/2 following equation 1.6 in Colella & Woodward, 1984: JCP 54, 174-201.
- Parameters
[in] hkm1: Width of cell k-1[in] hk: Width of cell k[in] hkp1: Width of cell k+1[in] hkp2: Width of cell k+2[in] ak: Average scalar value of cell k[in] akp1: Average scalar value of cell k+1[in] pk: PLM slope for cell k[in] pkp1: PLM slope for cell k+1[in] h_neglect: A negligibly small thickness [H ~> m or kg m-2]
-
real function mom_neutral_diffusion::ppm_ave(xL xL, xR xR, aL aL, aR aR, aMean aMean) Returns the average of a PPM reconstruction between two fractional positions.
- Parameters
[in] xl: Fraction position of left bound (0,1)[in] xr: Fraction position of right bound (0,1)[in] al: Left edge scalar value, at x=0[in] ar: Right edge scalar value, at x=1[in] amean: Average scalar value of cell
-
real function mom_neutral_diffusion::signum(a a, x x) A true signum function that returns either -abs(a), when x<0; or abs(a) when x>0; or 0 when x=0.
- Parameters
[in] a: The magnitude argument[in] x: The sign (or zero) argument
-
subroutine
plm_diff(nk nk, h h, S S, c_method c_method, b_method b_method, diff diff)¶ Returns PLM slopes for a column where the slopes are the difference in value across each cell. The limiting follows equation 1.8 in Colella & Woodward, 1984: JCP 54, 174-201.
- Parameters
[in] nk: Number of levels[in] h: Layer thickness [H ~> m or kg m-2][in] s: Layer salinity (conc, e.g. ppt)[in] c_method: Method to use for the centered difference[in] b_method: =1, use PCM in first/last cell, =2 uses linear extrapolation[inout] diff: Scalar difference across layer (conc, e.g. ppt) determined by the following values for c_method:Second order finite difference (not recommended)
Second order finite volume (used in original PPM)
Finite-volume weighted least squares linear fit
-
real function mom_neutral_diffusion::fv_diff(hkm1 hkm1, hk hk, hkp1 hkp1, Skm1 Skm1, Sk Sk, Skp1 Skp1) Returns the cell-centered second-order finite volume (unlimited PLM) slope using three consecutive cell widths and average values. Slope is returned as a difference across the central cell (i.e. units of scalar S). Discretization follows equation 1.7 in Colella & Woodward, 1984: JCP 54, 174-201.
- Parameters
[in] hkm1: Left cell width[in] hk: Center cell width[in] hkp1: Right cell width[in] skm1: Left cell average value[in] sk: Center cell average value[in] skp1: Right cell average value
-
real function mom_neutral_diffusion::fvlsq_slope(hkm1 hkm1, hk hk, hkp1 hkp1, Skm1 Skm1, Sk Sk, Skp1 Skp1) Returns the cell-centered second-order weighted least squares slope using three consecutive cell widths and average values. Slope is returned as a gradient (i.e. units of scalar S over width units).
- Parameters
[in] hkm1: Left cell width[in] hk: Center cell width[in] hkp1: Right cell width[in] skm1: Left cell average value[in] sk: Center cell average value[in] skp1: Right cell average value
-
subroutine
find_neutral_surface_positions_continuous(nk nk, Pl Pl, Tl Tl, Sl Sl, dRdTl dRdTl, dRdSl dRdSl, Pr Pr, Tr Tr, Sr Sr, dRdTr dRdTr, dRdSr dRdSr, PoL PoL, PoR PoR, KoL KoL, KoR KoR, hEff hEff)¶ Returns positions within left/right columns of combined interfaces using continuous reconstructions of T/S.
- Parameters
[in] nk: Number of levels[in] pl: Left-column interface pressure [Pa][in] tl: Left-column interface potential temperature [degC][in] sl: Left-column interface salinity [ppt][in] drdtl: Left-column dRho/dT [kg m-3 degC-1][in] drdsl: Left-column dRho/dS [kg m-3 ppt-1][in] pr: Right-column interface pressure [Pa][in] tr: Right-column interface potential temperature [degC][in] sr: Right-column interface salinity [ppt][in] drdtr: Left-column dRho/dT [kg m-3 degC-1][in] drdsr: Left-column dRho/dS [kg m-3 ppt-1][inout] pol: Fractional position of neutral surface within layer KoL of left column[inout] por: Fractional position of neutral surface within layer KoR of right column[inout] kol: Index of first left interface above neutral surface[inout] kor: Index of first right interface above neutral surface[inout] heff: Effective thickness between two neutral surfaces [Pa]
-
subroutine
find_neutral_surface_positions_discontinuous(CS CS, nk nk, ns ns, Pres_l Pres_l, hcol_l hcol_l, Tl Tl, Sl Sl, dRdT_l dRdT_l, dRdS_l dRdS_l, stable_l stable_l, Pres_r Pres_r, hcol_r hcol_r, Tr Tr, Sr Sr, dRdT_r dRdT_r, dRdS_r dRdS_r, stable_r stable_r, PoL PoL, PoR PoR, KoL KoL, KoR KoR, hEff hEff, ppoly_T_l ppoly_T_l, ppoly_S_l ppoly_S_l, ppoly_T_r ppoly_T_r, ppoly_S_r ppoly_S_r)¶ Higher order version of find_neutral_surface_positions. Returns positions within left/right columns of combined interfaces using intracell reconstructions of T/S.
- Parameters
[inout] cs: Neutral diffusion control structure[in] nk: Number of levels[in] ns: Number of neutral surfaces[in] pres_l: Left-column interface pressure [Pa][in] hcol_l: Left-column layer thicknesses[in] tl: Left-column top interface potential temperature [degC][in] sl: Left-column top interface salinity [ppt][in] drdt_l: Left-column, top interface dRho/dT [kg m-3 degC-1][in] drds_l: Left-column, top interface dRho/dS [kg m-3 ppt-1][in] stable_l: Left-column, top interface is stable[in] pres_r: Right-column interface pressure [Pa][in] hcol_r: Left-column layer thicknesses[in] tr: Right-column top interface potential temperature [degC][in] sr: Right-column top interface salinity [ppt][in] drdt_r: Right-column, top interface dRho/dT [kg m-3 degC-1][in] drds_r: Right-column, top interface dRho/dS [kg m-3 ppt-1][in] stable_r: Right-column, top interface is stable[inout] pol: Fractional position of neutral surface within layer KoL of left column[inout] por: Fractional position of neutral surface within layer KoR of right column[inout] kol: Index of first left interface above neutral surface[inout] kor: Index of first right interface above neutral surface[inout] heff: Effective thickness between two neutral surfaces [Pa][in] ppoly_t_l: Left-column coefficients of T reconstruction[in] ppoly_s_l: Left-column coefficients of S reconstruction[in] ppoly_t_r: Right-column coefficients of T reconstruction[in] ppoly_s_r: Right-column coefficients of S reconstruction
-
real function mom_neutral_diffusion::absolute_position(n n, ns ns, Pint Pint, Karr Karr, NParr NParr, k_surface k_surface) Converts non-dimensional position within a layer to absolute position (for debugging)
- Parameters
[in] n: Number of levels[in] ns: Number of neutral surfaces[in] pint: Position of interfaces [Pa][in] karr: Index of interface above position[in] nparr: Non-dimensional position within layer Karr(:)[in] k_surface: k-interface to query
-
real function, dimension(ns) mom_neutral_diffusion::absolute_positions(n n, ns ns, Pint Pint, Karr Karr, NParr NParr) Converts non-dimensional positions within layers to absolute positions (for debugging)
- Parameters
[in] n: Number of levels[in] ns: Number of neutral surfaces[in] pint: Position of interface [Pa][in] karr: Indexes of interfaces about positions[in] nparr: Non-dimensional positions within layers Karr(:)
-
subroutine
neutral_surface_flux(nk nk, nsurf nsurf, deg deg, hl hl, hr hr, Tl Tl, Tr Tr, PiL PiL, PiR PiR, KoL KoL, KoR KoR, hEff hEff, Flx Flx, continuous continuous, h_neglect h_neglect, remap_CS remap_CS, h_neglect_edge h_neglect_edge)¶ Returns a single column of neutral diffusion fluxes of a tracer.
- Parameters
[in] nk: Number of levels[in] nsurf: Number of neutral surfaces[in] deg: Degree of polynomial reconstructions[in] hl: Left-column layer thickness [Pa][in] hr: Right-column layer thickness [Pa][in] tl: Left-column layer tracer (conc, e.g. degC)[in] tr: Right-column layer tracer (conc, e.g. degC)[in] pil: Fractional position of neutral surface within layer KoL of left column[in] pir: Fractional position of neutral surface within layer KoR of right column[in] kol: Index of first left interface above neutral surface[in] kor: Index of first right interface above neutral surface[in] heff: Effective thickness between two neutral surfaces [Pa][inout] flx: Flux of tracer between pairs of neutral layers (conc H)[in] continuous: True if using continuous reconstruction[in] h_neglect: A negligibly small width for the purpose of cell reconstructions in the same units as h0.[in] remap_cs: Remapping control structure used to create sublayers[in] h_neglect_edge: A negligibly small width for the purpose of edge value calculations in the same units as h0.
-
subroutine
neutral_surface_t_eval(nk nk, ns ns, k_sub k_sub, Ks Ks, Ps Ps, T_mean T_mean, T_int T_int, deg deg, iMethod iMethod, T_poly T_poly, T_top T_top, T_bot T_bot, T_sub T_sub, T_top_int T_top_int, T_bot_int T_bot_int, T_layer T_layer)¶ Evaluate various parts of the reconstructions to calculate gradient-based flux limter.
- Parameters
[in] nk: Number of cell everages[in] ns: Number of neutral surfaces[in] k_sub: Index of current neutral layer[in] ks: List of the layers associated with each neutral surface[in] ps: List of the positions within a layer of each surface[in] t_mean: Cell average of tracer[in] t_int: Cell interface values of tracer from reconstruction[in] deg: Degree of reconstruction polynomial (e.g. 1 is linear)[in] imethod: Method of integration to use[in] t_poly: Coefficients of polynomial reconstructions[out] t_top: Tracer value at top (across discontinuity if necessary)[out] t_bot: Tracer value at bottom (across discontinuity if necessary)[out] t_sub: Average of the tracer value over the sublayer[out] t_top_int: Tracer value at top interface of neutral layer[out] t_bot_int: Tracer value at bottom interface of neutral layer[out] t_layer: Cell-average that the the reconstruction belongs to
-
subroutine
ppm_left_right_edge_values(nk nk, Tl Tl, Ti Ti, aL aL, aR aR)¶ Discontinuous PPM reconstructions of the left/right edge values within a cell.
- Parameters
[in] nk: Number of levels[in] tl: Layer tracer (conc, e.g. degC)[in] ti: Interface tracer (conc, e.g. degC)[inout] al: Left edge value of tracer (conc, e.g. degC)[inout] ar: Right edge value of tracer (conc, e.g. degC)
-
logical function, public mom_neutral_diffusion::neutral_diffusion_unit_tests(verbose verbose) Returns true if unit tests of neutral_diffusion functions fail. Otherwise returns false.
- Parameters
[in] verbose: If true, write results to stdout
-
logical function mom_neutral_diffusion::ndiff_unit_tests_continuous(verbose verbose) Returns true if unit tests of neutral_diffusion functions fail. Otherwise returns false.
- Parameters
[in] verbose: If true, write results to stdout
-
logical function mom_neutral_diffusion::ndiff_unit_tests_discontinuous(verbose verbose) - Parameters
[in] verbose: It true, write results to stdout
-
logical function mom_neutral_diffusion::test_fv_diff(verbose verbose, hkm1 hkm1, hk hk, hkp1 hkp1, Skm1 Skm1, Sk Sk, Skp1 Skp1, Ptrue Ptrue, title title) Returns true if a test of fv_diff() fails, and conditionally writes results to stream.
- Parameters
[in] verbose: If true, write results to stdout[in] hkm1: Left cell width[in] hk: Center cell width[in] hkp1: Right cell width[in] skm1: Left cell average value[in] sk: Center cell average value[in] skp1: Right cell average value[in] ptrue: True answer [Pa][in] title: Title for messages
-
logical function mom_neutral_diffusion::test_fvlsq_slope(verbose verbose, hkm1 hkm1, hk hk, hkp1 hkp1, Skm1 Skm1, Sk Sk, Skp1 Skp1, Ptrue Ptrue, title title) Returns true if a test of fvlsq_slope() fails, and conditionally writes results to stream.
- Parameters
[in] verbose: If true, write results to stdout[in] hkm1: Left cell width[in] hk: Center cell width[in] hkp1: Right cell width[in] skm1: Left cell average value[in] sk: Center cell average value[in] skp1: Right cell average value[in] ptrue: True answer [Pa][in] title: Title for messages
-
logical function mom_neutral_diffusion::test_ifndp(verbose verbose, rhoNeg rhoNeg, Pneg Pneg, rhoPos rhoPos, Ppos Ppos, Ptrue Ptrue, title title) Returns true if a test of interpolate_for_nondim_position() fails, and conditionally writes results to stream.
- Parameters
[in] verbose: If true, write results to stdout[in] rhoneg: Lighter density [kg m-3][in] pneg: Interface position of lighter density [Pa][in] rhopos: Heavier density [kg m-3][in] ppos: Interface position of heavier density [Pa][in] ptrue: True answer [Pa][in] title: Title for messages
-
logical function mom_neutral_diffusion::test_data1d(verbose verbose, nk nk, Po Po, Ptrue Ptrue, title title) Returns true if comparison of Po and Ptrue fails, and conditionally writes results to stream.
- Parameters
[in] verbose: If true, write results to stdout[in] nk: Number of layers[in] po: Calculated answer[in] ptrue: True answer[in] title: Title for messages
-
logical function mom_neutral_diffusion::test_data1di(verbose verbose, nk nk, Po Po, Ptrue Ptrue, title title) Returns true if comparison of Po and Ptrue fails, and conditionally writes results to stream.
- Parameters
[in] verbose: If true, write results to stdout[in] nk: Number of layers[in] po: Calculated answer[in] ptrue: True answer[in] title: Title for messages
-
logical function mom_neutral_diffusion::test_nsp(verbose verbose, ns ns, KoL KoL, KoR KoR, pL pL, pR pR, hEff hEff, KoL0 KoL0, KoR0 KoR0, pL0 pL0, pR0 pR0, hEff0 hEff0, title title) Returns true if output of find_neutral_surface_positions() does not match correct values, and conditionally writes results to stream.
- Parameters
[in] verbose: If true, write results to stdout[in] ns: Number of surfaces[in] kol: Index of first left interface above neutral surface[in] kor: Index of first right interface above neutral surface[in] pl: Fractional position of neutral surface within layer KoL of left column[in] pr: Fractional position of neutral surface within layer KoR of right column[in] heff: Effective thickness between two neutral surfaces [Pa][in] kol0: Correct value for KoL[in] kor0: Correct value for KoR[in] pl0: Correct value for pL[in] pr0: Correct value for pR[in] heff0: Correct value for hEff[in] title: Title for messages
-
logical function mom_neutral_diffusion::compare_nsp_row(KoL KoL, KoR KoR, pL pL, pR pR, KoL0 KoL0, KoR0 KoR0, pL0 pL0, pR0 pR0) Compares a single row, k, of output from find_neutral_surface_positions()
- Parameters
[in] kol: Index of first left interface above neutral surface[in] kor: Index of first right interface above neutral surface[in] pl: Fractional position of neutral surface within layer KoL of left column[in] pr: Fractional position of neutral surface within layer KoR of right column[in] kol0: Correct value for KoL[in] kor0: Correct value for KoR[in] pl0: Correct value for pL[in] pr0: Correct value for pR
-
logical function mom_neutral_diffusion::test_rnp(expected_pos expected_pos, test_pos test_pos, title title) Compares output position from refine_nondim_position with an expected value.
- Parameters
[in] expected_pos: The expected position[in] test_pos: The position returned by the code[in] title: A label for this test
-
subroutine, public mom_neutral_diffusion::neutral_diffusion_end(CS CS) Deallocates neutral_diffusion control structure.
- Parameters
cs: Neutral diffusion control structure
Variables
-
character(len=40) mom_neutral_diffusion::mdl = "MOM_neutral_diffusion" module name
-
-
namespace
mom_neutral_diffusion_aux¶ A column-wise toolbox for implementing neutral diffusion.
Functions
-
subroutine, public mom_neutral_diffusion_aux::set_ndiff_aux_params(CS CS, deg deg, max_iter max_iter, drho_tol drho_tol, xtol xtol, ref_pres ref_pres, force_brent force_brent, EOS EOS, debug debug) Initialize the parameters used to iteratively find the neutral direction.
- Parameters
[inout] cs: Control structure for refine_pos[in] deg: Degree of polynommial used in reconstruction[in] max_iter: Maximum number of iterations[in] drho_tol: Tolerance for function convergence[in] xtol: Tolerance for change in position[in] ref_pres: Reference pressure to use[in] force_brent: Force Brent method for linear, TEOS-10, and WRIGHT[in] debug: If true, print output use to help debug neutral diffusion[in] eos: Equation of state
-
subroutine, public mom_neutral_diffusion_aux::mark_unstable_cells(nk nk, dRdT dRdT, dRdS dRdS, T T, S S, stable_cell stable_cell, ns ns) Given the reconsturcitons of dRdT, dRdS, T, S mark the cells which are stably stratified parts of the water column For an layer to be unstable the top interface must be denser than the bottom or the bottom interface of the layer.
- Parameters
[in] nk: Number of levels in a column[in] drdt: drho/dT [kg m-3 degC-1] at interfaces[in] drds: drho/dS [kg m-3 ppt-1] at interfaces[in] t: Temperature [degC] at interfaces[in] s: Salinity [ppt] at interfaces[out] stable_cell: True if this cell is unstably stratified[out] ns: Number of neutral surfaces in unmasked part of the column
-
subroutine, public mom_neutral_diffusion_aux::increment_interface(nk nk, kl kl, ki ki, stable stable, reached_bottom reached_bottom, searching_this_column searching_this_column, searching_other_column searching_other_column) Increments the interface which was just connected and also set flags if the bottom is reached.
- Parameters
[in] nk: Number of vertical levels[inout] kl: Current layer (potentially updated)[inout] ki: Current interface[in] stable: True if the cell is stably stratified[inout] reached_bottom: Updated when kl == nk and ki == 2[inout] searching_this_column: Updated when kl == nk and ki == 2[inout] searching_other_column: Updated when kl == nk and ki == 2
-
real function, public mom_neutral_diffusion_aux::calc_drho(T1 T1, S1 S1, dRdT1 dRdT1, dRdS1 dRdS1, T2 T2, S2 S2, dRdT2 dRdT2, dRdS2 dRdS2) Calculates difference in density at two points (rho1-rho2) with known density derivatives, T, and S.
- Parameters
[in] t1: Temperature at point 1[in] s1: Salinity at point 1[in] drdt1: dRhodT at point 1[in] drds1: dRhodS at point 1[in] t2: Temperature at point 2[in] s2: Salinity at point 2[in] drdt2: dRhodT at point 2[in] drds2: dRhodS at point
-
subroutine, public mom_neutral_diffusion_aux::drho_at_pos(CS CS, T_ref T_ref, S_ref S_ref, alpha_ref alpha_ref, beta_ref beta_ref, P_top P_top, P_bot P_bot, ppoly_T ppoly_T, ppoly_S ppoly_S, x0 x0, delta_rho delta_rho, P_out P_out, T_out T_out, S_out S_out, alpha_avg_out alpha_avg_out, beta_avg_out beta_avg_out, delta_T_out delta_T_out, delta_S_out delta_S_out) Calculate the difference in neutral density between a reference T, S, alpha, and beta at a point on the polynomial reconstructions of T, S.
- Parameters
[in] cs: Control structure with parameters for this module[in] t_ref: Temperature at reference surface[in] s_ref: Salinity at reference surface[in] alpha_ref: dRho/dT at reference surface[in] beta_ref: dRho/dS at reference surface[in] p_top: Pressure (Pa) at top interface of layer to be searched[in] p_bot: Pressure (Pa) at bottom interface[in] ppoly_t: Coefficients of T reconstruction[in] ppoly_s: Coefficients of S reconstruciton[in] x0: Nondimensional position to evaluate[out] delta_rho: The density difference from a reference value[out] p_out: Pressure at point x0[out] t_out: Temperature at point x0[out] s_out: Salinity at point x0[out] alpha_avg_out: Average of alpha between reference and x0[out] beta_avg_out: Average of beta between reference and x0[out] delta_t_out: Difference in temperature between reference and x0[out] delta_s_out: Difference in salinity between reference and x0
-
subroutine, public mom_neutral_diffusion_aux::search_other_column(dRhoTop dRhoTop, dRhoBot dRhoBot, Ptop Ptop, Pbot Pbot, lastP lastP, lastK lastK, kl kl, kl_0 kl_0, ki ki, top_connected top_connected, bot_connected bot_connected, out_P out_P, out_K out_K, search_layer search_layer) Searches the “other” (searched) column for the position of the neutral surface.
- Parameters
[in] drhotop: Density difference across top interface[in] drhobot: Density difference across top interface[in] ptop: Pressure at top interface[in] pbot: Pressure at bottom interface[in] lastp: Last position connected in the searched column[in] lastk: Last layer connected in the searched column[in] kl: Layer in the searched column[in] kl_0: Layer in the searched column[in] ki: Interface of the searched column[inout] top_connected: True if the top interface was pointed to[inout] bot_connected: True if the top interface was pointed to[out] out_p: Position within searched column[out] out_k: Layer within searched column[out] search_layer: Neutral surface within cell
-
real function, public mom_neutral_diffusion_aux::interpolate_for_nondim_position(dRhoNeg dRhoNeg, Pneg Pneg, dRhoPos dRhoPos, Ppos Ppos) Returns the non-dimensional position between Pneg and Ppos where the interpolated density difference equals zero. The result is always bounded to be between 0 and 1.
- Parameters
[in] drhoneg: Negative density difference[in] pneg: Position of negative density difference[in] drhopos: Positive density difference[in] ppos: Position of positive density difference
-
real function, public mom_neutral_diffusion_aux::refine_nondim_position(CS CS, T_ref T_ref, S_ref S_ref, alpha_ref alpha_ref, beta_ref beta_ref, P_top P_top, P_bot P_bot, ppoly_T ppoly_T, ppoly_S ppoly_S, drho_top drho_top, drho_bot drho_bot, min_bound min_bound) Use root-finding methods to find where dRho = 0, based on the equation of state and the polynomial reconstructions of temperature, salinity. Initial guess is based on the zero crossing of based on linear profiles of dRho, T, and S, between the top and bottom interface. If second derivatives of the EOS are available, it starts with a Newton’s method. However, Newton’s method is not guaranteed to be bracketed, a check is performed to see if it it diverges outside the interval. In that case (or in the case that second derivatives are not available), Brent’s method is used following the implementation found at https://people.sc.fsu.edu/~jburkardt/f_src/brent/brent.f90.
- Parameters
[in] cs: Control structure with parameters for this module[in] t_ref: Temperature of the neutral surface at the searched from interface[in] s_ref: Salinity of the neutral surface at the searched from interface[in] alpha_ref: dRho/dT of the neutral surface at the searched from interface[in] beta_ref: dRho/dS of the neutral surface at the searched from interface[in] p_top: Pressure at the top interface in the layer to be searched[in] p_bot: Pressure at the bottom interface in the layer to be searched[in] ppoly_t: Coefficients of the order N polynomial reconstruction of T within the layer to be searched.[in] ppoly_s: Coefficients of the order N polynomial reconstruction of T within the layer to be searched.[in] drho_top: Delta rho at top interface (or previous position in cell[in] drho_bot: Delta rho at bottom interface[in] min_bound: Lower bound of position, also serves as the initial guess
-
subroutine, public mom_neutral_diffusion_aux::check_neutral_positions(CS CS, Ptop_l Ptop_l, Pbot_l Pbot_l, Ptop_r Ptop_r, Pbot_r Pbot_r, PoL PoL, PoR PoR, Tl_coeffs Tl_coeffs, Tr_coeffs Tr_coeffs, Sl_coeffs Sl_coeffs, Sr_coeffs Sr_coeffs) - Parameters
[in] cs: Control structure with parameters for this module[in] ptop_l: Pressure at top interface of left cell[in] pbot_l: Pressure at bottom interface of left cell[in] ptop_r: Pressure at top interface of right cell[in] pbot_r: Pressure at bottom interface of right cell[in] pol: Nondim position in left cell[in] por: Nondim position in right cell[in] tl_coeffs: T polynomial coefficients of left cell[in] tr_coeffs: T polynomial coefficients of right cell[in] sl_coeffs: S polynomial coefficients of left cell[in] sr_coeffs: S polynomial coefficients of right cell
-
subroutine, public mom_neutral_diffusion_aux::kahan_sum(sum sum, summand summand, c c) Do a compensated sum to account for roundoff level.
- Parameters
[inout] sum: Running sum[in] summand: Term to be added[inout] c: Keep track of roundoff
-
-
namespace
mom_obsolete_diagnostics¶ Provides a mechanism for recording diagnostic variables that are no longer valid, along with their replacement name if appropriate.
Functions
-
subroutine, public mom_obsolete_diagnostics::register_obsolete_diagnostics(param_file param_file, diag diag) Scan through the diag_table searching for obsolete parameters and issue informational messages and optionallly a FATAL error.
- Parameters
[in] param_file: The parameter file handle.[in] diag: A structure used to control diagnostics.
-
logical function mom_obsolete_diagnostics::found_in_diagtable(diag diag, varName varName, newVarName newVarName) Fakes a register of a diagnostic to find out if an obsolete parameter appears in the diag_table.
- Parameters
[in] diag: A structure used to control diagnostics.[in] varname: The obsolete diagnostic name[in] newvarname: The valid name of this diagnostic
-
-
namespace
mom_obsolete_params¶ Methods for testing for, and list of, obsolete run-time parameters.
Functions
-
subroutine, public mom_obsolete_params::find_obsolete_params(param_file param_file) Scans input parameter file for list obsolete parameters.
- Parameters
[in] param_file: Structure containing parameter file data.
-
subroutine, public mom_obsolete_params::obsolete_logical(param_file param_file, varname varname, warning_val warning_val, hint hint) Test for presence of obsolete LOGICAL in parameter file.
- Parameters
[in] param_file: Structure containing parameter file data.[in] varname: Name of obsolete LOGICAL parameter.[in] warning_val: An allowed value that causes a warning instead of an error.[in] hint: A hint to the user about what to do.
-
subroutine, public mom_obsolete_params::obsolete_char(param_file param_file, varname varname, hint hint) Test for presence of obsolete STRING in parameter file.
- Parameters
[in] param_file: Structure containing parameter file data.[in] varname: Name of obsolete STRING parameter.[in] hint: A hint to the user about what to do.
-
subroutine, public mom_obsolete_params::obsolete_real(param_file param_file, varname varname, warning_val warning_val, hint hint) Test for presence of obsolete REAL in parameter file.
- Parameters
[in] param_file: Structure containing parameter file data.[in] varname: Name of obsolete REAL parameter.[in] warning_val: An allowed value that causes a warning instead of an error.[in] hint: A hint to the user about what to do.
-
subroutine, public mom_obsolete_params::obsolete_int(param_file param_file, varname varname, warning_val warning_val, hint hint) Test for presence of obsolete INTEGER in parameter file.
- Parameters
[in] param_file: Structure containing parameter file data.[in] varname: Name of obsolete INTEGER parameter.[in] warning_val: An allowed value that causes a warning instead of an error.[in] hint: A hint to the user about what to do.
-
-
namespace
mom_ocmip2_cfc¶ Simulates CFCs using the OCMIP2 protocols.
By Robert Hallberg, 2007
This module contains the code that is needed to set up and use CFC-11 and CFC-12 in a fully coupled or ice-ocean model context using the OCMIP2 protocols
Unnamed Group
-
logical function, public mom_ocmip2_cfc::register_ocmip2_cfc(HI HI, GV GV, param_file param_file, CS CS, tr_Reg tr_Reg, restart_CS restart_CS) Register the OCMIP2 CFC tracers to be used with MOM and read the parameters that are used with this tracer package.
- Parameters
[in] hi: A horizontal index type structure.[in] gv: The ocean’s vertical grid structure.[in] param_file: A structure to parse for run-time parameters.cs: A pointer that is set to point to the control structure for this module.tr_reg: A pointer to the tracer registry.restart_cs: A pointer to the restart control structure.
-
subroutine, public mom_ocmip2_cfc::flux_init_ocmip2_cfc(CS CS, verbosity verbosity) This subroutine initializes the air-sea CFC fluxes, and optionally returns the indicies of these fluxes. It can safely be called multiple times.
- Parameters
cs: An optional pointer to the control structure for this module; if not present, the flux indicies are not stored.[in] verbosity: A 0-9 integer indicating a level of verbosity.
-
subroutine, public mom_ocmip2_cfc::initialize_ocmip2_cfc(restart restart, day day, G G, GV GV, US US, h h, diag diag, OBC OBC, CS CS, sponge_CSp sponge_CSp) Initialize the OCMP2 CFC tracer fields and set up the tracer output.
- Parameters
[in] restart: .true. if the fields have already been read from a restart file.[in] day: Time of the start of the run.[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] h: Layer thicknesses [H ~> m or kg m-2].[in] diag: A structure that is used to regulate diagnostic output.obc: This open boundary condition type specifies whether, where, and what open boundary conditions are used.cs: The control structure returned by a previous call to register_OCMIP2_CFC.sponge_csp: A pointer to the control structure for the sponges, if they are in use. Otherwise this may be unassociated.
-
subroutine
init_tracer_cfc(h h, tr tr, name name, land_val land_val, IC_val IC_val, G G, US US, CS CS)¶ This subroutine initializes a tracer array.
- Parameters
[in] g: The ocean’s grid structure[in] us: A dimensional unit scaling type[in] h: Layer thicknesses [H ~> m or kg m-2][out] tr: The tracer concentration array[in] name: The tracer name[in] land_val: A value the tracer takes over land[in] ic_val: The initial condition value for the tracercs: The control structure returned by a previous call to register_OCMIP2_CFC.
-
subroutine, public mom_ocmip2_cfc::ocmip2_cfc_column_physics(h_old h_old, h_new h_new, ea ea, eb eb, fluxes fluxes, dt dt, G G, GV GV, CS CS, evap_CFL_limit evap_CFL_limit, minimum_forcing_depth minimum_forcing_depth) This subroutine applies diapycnal diffusion, souces and sinks and any other column tracer physics or chemistry to the OCMIP2 CFC tracers. CFCs are relatively simple, as they are passive tracers with only a surface flux as a source.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h_old: Layer thickness before entrainment [H ~> m or kg m-2].[in] h_new: Layer thickness after entrainment [H ~> m or kg m-2].[in] ea: an array to which the amount of fluid entrained[in] eb: an array to which the amount of fluid entrained[in] fluxes: A structure containing pointers to thermodynamic and tracer forcing fields. Unused fields have NULL ptrs.[in] dt: The amount of time covered by this call [s]cs: The control structure returned by a previous call to register_OCMIP2_CFC.[in] evap_cfl_limit: Limit on the fraction of the water that can be fluxed out of the top layer in a timestep [nondim][in] minimum_forcing_depth: The smallest depth over which fluxes can be applied [m]
-
integer function, public mom_ocmip2_cfc::ocmip2_cfc_stock(h h, stocks stocks, G G, GV GV, CS CS, names names, units units, stock_index stock_index) This function calculates the mass-weighted integral of all tracer stocks, returning the number of stocks it has calculated. If the stock_index is present, only the stock corresponding to that coded index is returned.
- Return
The number of stocks calculated here.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] h: Layer thicknesses [H ~> m or kg m-2].[out] stocks: the mass-weighted integrated amount of each tracer, in kg times concentration units [kg conc].cs: The control structure returned by a previous call to register_OCMIP2_CFC.[out] names: The names of the stocks calculated.[out] units: The units of the stocks calculated.[in] stock_index: The coded index of a specific stock being sought.
-
subroutine, public mom_ocmip2_cfc::ocmip2_cfc_surface_state(state state, h h, G G, CS CS) This subroutine extracts the surface CFC concentrations and other fields that are shared with the atmosphere to calculate CFC fluxes.
- Parameters
[in] g: The ocean’s grid structure.[inout] state: A structure containing fields that describe the surface state of the ocean.[in] h: Layer thickness [H ~> m or kg m-2].cs: The control structure returned by a previous call to register_OCMIP2_CFC.
-
subroutine, public mom_ocmip2_cfc::ocmip2_cfc_end(CS CS) Deallocate any memory associated with the OCMIP2 CFC tracer package.
- Parameters
cs: The control structure returned by a previous call to register_OCMIP2_CFC.
Variables
-
integer, parameter mom_ocmip2_cfc::ntr = 2 the number of tracers in this module.
-
-
namespace
MOM_oda_driver_mod¶
-
namespace
mom_oda_driver_mod¶ Interfaces for MOM6 ensembles and data assimilation.
Unnamed Group
-
integer, parameter mom_oda_driver_mod::no_assim = 0 DA parameters.
-
integer, parameter mom_oda_driver_mod::oi_assim =1 DA parameters.
-
integer, parameter mom_oda_driver_mod::eakf_assim =2 DA parameters.
-
subroutine, public mom_oda_driver_mod::init_oda(Time Time, G G, GV GV, CS CS) initialize First_guess (prior) and Analysis grid information for all ensemble members
- Parameters
[in] time: The current model time.g: domain and grid information for ocean model[in] gv: The ocean’s vertical grid structure[inout] cs: The DA control structure
-
subroutine, public mom_oda_driver_mod::set_prior_tracer(Time Time, G G, GV GV, h h, tv tv, CS CS) Copy ensemble member tracers to ensemble vector.
- Parameters
[in] time: The current model timeg: domain and grid information for ocean model[in] gv: The ocean’s vertical grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][in] tv: A structure pointing to various thermodynamic variablescs: ocean DA control structure
-
subroutine, public mom_oda_driver_mod::get_posterior_tracer(Time Time, CS CS, h h, tv tv, increment increment) Returns posterior adjustments or full state Note that only those PEs associated with an ensemble member receive data.
- Parameters
[in] time: the current model timecs: ocean DA control structureh: Layer thicknesses [H ~> m or kg m-2]tv: A structure pointing to various thermodynamic variables[in] increment: True if returning increment only
-
subroutine, public mom_oda_driver_mod::oda(Time Time, CS CS) Gather observations and sall ODA routines.
- Parameters
[in] time: the current model time[inout] cs: the ocean DA control structure
-
subroutine, public mom_oda_driver_mod::oda_end(CS CS) Finalize DA module.
- Parameters
[inout] cs: the ocean DA control structure
-
subroutine
init_ocean_ensemble(CS CS, Grid Grid, GV GV, ens_size ens_size)¶ Initialize DA module.
- Parameters
cs: Pointer to ODA control structuregrid: Pointer to ocean analysis gridgv: Pointer to DA vertical grid[in] ens_size: ensemble size
-
subroutine, public mom_oda_driver_mod::set_analysis_time(Time Time, CS CS) Set the next analysis time.
- Parameters
[in] time: the current model time[inout] cs: the DA control structure
-
subroutine, public mom_oda_driver_mod::save_obs_diff(filename filename, CS CS) Write observation differences to a file.
- Parameters
[in] filename: name of output file[in] cs: pointer to DA control structure
-
subroutine, public mom_oda_driver_mod::apply_oda_tracer_increments(dt dt, G G, tv tv, h h, CS CS) Apply increments to tracers.
- Parameters
[in] dt: The tracer timestep [s][in] g: ocean grid structure[inout] tv: A structure pointing to various thermodynamic variables[in] h: layer thickness [H ~> m or kg m-2][inout] cs: the data assimilation structure
-
-
namespace
mom_offline_aux¶ Contains routines related to offline transport of tracers. These routines are likely to be called from the MOM_offline_main module.
Functions
-
subroutine, public mom_offline_aux::update_h_horizontal_flux(G G, GV GV, uhtr uhtr, vhtr vhtr, h_pre h_pre, h_new h_new) This updates thickness based on the convergence of horizontal mass fluxes NOTE: Only used in non-ALE mode.
- Parameters
g: ocean grid structuregv: ocean vertical grid structure[in] uhtr: Accumulated mass flux through zonal face [kg][in] vhtr: Accumulated mass flux through meridional face [kg][in] h_pre: Previous layer thicknesses [kg m-2].[inout] h_new: Updated layer thicknesses [kg m-2].
-
subroutine, public mom_offline_aux::update_h_vertical_flux(G G, GV GV, ea ea, eb eb, h_pre h_pre, h_new h_new) Updates layer thicknesses due to vertical mass transports NOTE: Only used in non-ALE configuration.
- Parameters
g: ocean grid structuregv: ocean vertical grid structure[in] ea: Mass of fluid entrained from the layer[in] eb: Mass of fluid entrained from the layer[in] h_pre: Layer thicknesses at the end of the previous[inout] h_new: Updated layer thicknesses [kg m-2].
-
subroutine, public mom_offline_aux::limit_mass_flux_3d(G G, GV GV, uh uh, vh vh, ea ea, eb eb, h_pre h_pre) This routine limits the mass fluxes so that the a layer cannot be completely depleted. NOTE: Only used in non-ALE mode.
- Parameters
g: ocean grid structuregv: ocean vertical grid structure[inout] uh: Mass flux through zonal face [kg][inout] vh: Mass flux through meridional face [kg][inout] ea: Mass of fluid entrained from the layer[inout] eb: Mass of fluid entrained from the layer[in] h_pre: Layer thicknesses at the end of the previous
-
subroutine, public mom_offline_aux::distribute_residual_uh_barotropic(G G, GV GV, hvol hvol, uh uh) In the case where offline advection has failed to converge, redistribute the u-flux into remainder of the water column as a barotropic equivalent.
- Parameters
g: ocean grid structuregv: ocean vertical grid structure[in] hvol: Mass of water in the cells at the end[inout] uh: Zonal mass transport within a timestep [kg]
-
subroutine, public mom_offline_aux::distribute_residual_vh_barotropic(G G, GV GV, hvol hvol, vh vh) Redistribute the v-flux as a barotropic equivalent.
- Parameters
g: ocean grid structuregv: ocean vertical grid structure[in] hvol: Mass of water in the cells at the end[inout] vh: Meridional mass transport within a timestep [kg]
-
subroutine, public mom_offline_aux::distribute_residual_uh_upwards(G G, GV GV, hvol hvol, uh uh) In the case where offline advection has failed to converge, redistribute the u-flux into layers above.
- Parameters
g: ocean grid structuregv: ocean vertical grid structure[in] hvol: Mass of water in the cells at the end[inout] uh: Zonal mass transport within a timestep [kg]
-
subroutine, public mom_offline_aux::distribute_residual_vh_upwards(G G, GV GV, hvol hvol, vh vh) In the case where offline advection has failed to converge, redistribute the u-flux into layers above.
- Parameters
g: ocean grid structuregv: ocean vertical grid structure[in] hvol: Mass of water in the cells at the end[inout] vh: Meridional mass transport within a timestep [kg]
-
subroutine, public mom_offline_aux::offline_add_diurnal_sw(fluxes fluxes, G G, Time_start Time_start, Time_end Time_end) add_diurnal_SW adjusts the shortwave fluxes in an forcying_type variable to add a synthetic diurnal cycle. Adapted from SIS2
- Parameters
[inout] fluxes: The type with atmospheric fluxes to be adjusted.[in] g: The ocean lateral grid type.[in] time_start: The start time for this step.[in] time_end: The ending time for this step.
-
subroutine, public mom_offline_aux::update_offline_from_files(G G, GV GV, nk_input nk_input, mean_file mean_file, sum_file sum_file, snap_file snap_file, surf_file surf_file, h_end h_end, uhtr uhtr, vhtr vhtr, temp_mean temp_mean, salt_mean salt_mean, mld mld, Kd Kd, fluxes fluxes, ridx_sum ridx_sum, ridx_snap ridx_snap, read_mld read_mld, read_sw read_sw, read_ts_uvh read_ts_uvh, do_ale_in do_ale_in) Controls the reading in 3d mass fluxes, diffusive fluxes, and other fields stored in a previous integration of the online model.
- Parameters
[inout] g: Horizontal grid type[in] gv: Vertical grid type[in] nk_input: Number of levels in input file[in] mean_file: Name of file with averages fields[in] sum_file: Name of file with summed fields[in] snap_file: Name of file with snapshot fields[in] surf_file: Name of file with surface fields[inout] uhtr: Zonal mass fluxes [kg][inout] vhtr: Meridional mass fluxes [kg][inout] h_end: End of timestep layer thickness[inout] temp_mean: Averaged temperature[inout] salt_mean: Averaged salinity[inout] mld: Averaged mixed layer depth[inout] kd: Diapycnal diffusivities at interfaces[inout] fluxes: Fields with surface fluxes[in] ridx_sum: Read index for sum, mean, and surf files[in] ridx_snap: Read index for snapshot file[in] read_mld: True if reading in MLD[in] read_sw: True if reading in radiative fluxes[in] read_ts_uvh: True if reading in uh, vh, and h[in] do_ale_in: True if using ALE algorithms
-
subroutine, public mom_offline_aux::update_offline_from_arrays(G G, GV GV, nk_input nk_input, ridx_sum ridx_sum, mean_file mean_file, sum_file sum_file, snap_file snap_file, uhtr uhtr, vhtr vhtr, hend hend, uhtr_all uhtr_all, vhtr_all vhtr_all, hend_all hend_all, temp temp, salt salt, temp_all temp_all, salt_all salt_all) Fields for offline transport are copied from the stored arrays read during initialization.
- Parameters
[inout] g: Horizontal grid type[in] gv: Vertical grid type[in] nk_input: Number of levels in input file[in] ridx_sum: Index to read from[in] mean_file: Name of file with averages fields[in] sum_file: Name of file with summed fields[in] snap_file: Name of file with snapshot fields[inout] uhtr: Zonal mass fluxes [kg][inout] vhtr: Meridional mass fluxes [kg][inout] hend: End of timestep layer thickness [kg m-2][inout] uhtr_all: Zonal mass fluxes [kg][inout] vhtr_all: Meridional mass fluxes [kg][inout] hend_all: End of timestep layer thickness [kg m-2][inout] temp: Temperature array[inout] salt: Salinity array[inout] temp_all: Temperature array[inout] salt_all: Salinity array
-
integer function, public mom_offline_aux::next_modulo_time(inidx inidx, numtime numtime) Calculates the next timelevel to read from the input fields. This allows the ‘looping’ of the fields.
-
-
namespace
mom_offline_main¶ The routines here implement the offline tracer algorithm used in MOM6. These are called from step_offline Some routines called here can be found in the MOM_offline_aux module.
Unnamed Group
-
subroutine, public mom_offline_main::offline_advection_ale(fluxes fluxes, Time_start Time_start, time_interval time_interval, CS CS, id_clock_ale id_clock_ale, h_pre h_pre, uhtr uhtr, vhtr vhtr, converged converged) 3D advection is done by doing flux-limited nonlinear horizontal advection interspersed with an ALE regridding/remapping step. The loop in this routine is exited if remaining residual transports are below a runtime-specified value or a maximum number of iterations is reached.
- Parameters
[inout] fluxes: pointers to forcing fields[in] time_start: starting time of a segment, as a time type[in] time_interval: time intervalcs: control structure for offline module[in] id_clock_ale: Clock for ALE routines[inout] h_pre: layer thicknesses before advection[inout] uhtr: Zonal mass transport [H m2 ~> m3 or kg][inout] vhtr: Meridional mass transport [H m2 ~> m3 or kg][out] converged: True if the iterations have converged
-
subroutine, public mom_offline_main::offline_redistribute_residual(CS CS, h_pre h_pre, uhtr uhtr, vhtr vhtr, converged converged) In the case where the main advection routine did not converge, something needs to be done with the remaining transport. Two different ways are offered, ‘barotropic’ means that the residual is distributed equally throughout the water column. ‘upwards’ attempts to redistribute the transport in the layers above and will eventually work down the entire water column.
- Parameters
cs: control structure from initialize_MOM[inout] h_pre: layer thicknesses before advection[inout] uhtr: Zonal mass transport[inout] vhtr: Meridional mass transport[in] converged: True if the iterations have converged
-
real function mom_offline_main::remaining_transport_sum(CS CS, uhtr uhtr, vhtr vhtr) Sums any non-negligible remaining transport to check for advection convergence.
- Parameters
cs: control structure for offline module[in] uhtr: Zonal mass transport[in] vhtr: Meridional mass transport
-
subroutine, public mom_offline_main::offline_diabatic_ale(fluxes fluxes, Time_start Time_start, Time_end Time_end, CS CS, h_pre h_pre, eatr eatr, ebtr ebtr) The vertical/diabatic driver for offline tracers. First the eatr/ebtr associated with the interpolated vertical diffusivities are calculated and then any tracer column functions are done which can include vertical diffuvities and source/sink terms.
- Parameters
[inout] fluxes: pointers to forcing fields[in] time_start: starting time of a segment, as a time type[in] time_end: time intervalcs: control structure from initialize_MOM[inout] h_pre: layer thicknesses before advection [H ~> m or kg m-2][inout] eatr: Entrainment from layer above [H ~> m or kg m-2][inout] ebtr: Entrainment from layer below [H ~> m or kg m-2]
-
subroutine, public mom_offline_main::offline_fw_fluxes_into_ocean(G G, GV GV, CS CS, fluxes fluxes, h h, in_flux_optional in_flux_optional) Apply positive freshwater fluxes (into the ocean) and update netMassOut with only the negative (out of the ocean) fluxes.
- Parameters
[inout] cs: Offline control structure[in] g: Grid structure[in] gv: ocean vertical grid structure[inout] fluxes: Surface fluxes container[inout] h: Layer thickness [H ~> m or kg m-2][in] in_flux_optional: The total time-integrated amount
-
subroutine, public mom_offline_main::offline_fw_fluxes_out_ocean(G G, GV GV, CS CS, fluxes fluxes, h h, out_flux_optional out_flux_optional) Apply negative freshwater fluxes (out of the ocean)
- Parameters
[inout] cs: Offline control structure[in] g: Grid structure[in] gv: ocean vertical grid structure[inout] fluxes: Surface fluxes container[inout] h: Layer thickness [H ~> m or kg m-2][in] out_flux_optional: The total time-integrated amount
-
subroutine, public mom_offline_main::offline_advection_layer(fluxes fluxes, Time_start Time_start, time_interval time_interval, CS CS, h_pre h_pre, eatr eatr, ebtr ebtr, uhtr uhtr, vhtr vhtr) When in layer mode, 3D horizontal advection using stored mass fluxes must be used. Horizontal advection is done via tracer_advect, whereas the vertical component is actually handled by vertdiff in tracer_column_fns.
- Parameters
[inout] fluxes: pointers to forcing fields[in] time_start: starting time of a segment, as a time type[in] time_interval: Offline transport time intervalcs: Control structure for offline module[inout] h_pre: layer thicknesses before advection[inout] eatr: Entrainment from layer above[inout] ebtr: Entrainment from layer below[inout] uhtr: Zonal mass transport[inout] vhtr: Meridional mass transport
-
subroutine, public mom_offline_main::update_offline_fields(CS CS, h h, fluxes fluxes, do_ale do_ale) Update fields used in this round of offline transport. First fields are updated from files or from arrays read during initialization. Then if in an ALE-dependent coordinate, regrid/remap fields.
- Parameters
cs: Control structure for offline moduleh: The regridded layer thicknesses[inout] fluxes: Pointers to forcing fields[in] do_ale: True if using ALE
-
subroutine, public mom_offline_main::register_diags_offline_transport(Time Time, diag diag, CS CS) Initialize additional diagnostics required for offline tracer transport.
- Parameters
cs: Control structure for offline module[in] time: current model time[in] diag: Structure that regulates diagnostic output
-
subroutine, public mom_offline_main::post_offline_convergence_diags(CS CS, h_off h_off, h_end h_end, uhtr uhtr, vhtr vhtr) Posts diagnostics related to offline convergence diagnostics.
- Parameters
[in] cs: Offline control structure[inout] h_off: Thicknesses at end of offline step[inout] h_end: Stored thicknesses[inout] uhtr: Remaining zonal mass transport[inout] vhtr: Remaining meridional mass transport
-
subroutine, public mom_offline_main::extract_offline_main(CS CS, uhtr uhtr, vhtr vhtr, eatr eatr, ebtr ebtr, h_end h_end, accumulated_time accumulated_time, dt_offline dt_offline, dt_offline_vertical dt_offline_vertical, skip_diffusion skip_diffusion) Extracts members of the offline main control structure. All arguments are optional except the control structure itself.
- Parameters
[in] cs: Offline control structureuhtr: Remaining zonal mass transport [H m2 ~> m3 or kg]vhtr: Remaining meridional mass transport [H m2 ~> m3 or kg]eatr: Amount of fluid entrained from the layer above within one time step [H ~> m or kg m-2]ebtr: Amount of fluid entrained from the layer below within one time step [H ~> m or kg m-2]h_end: Thicknesses at the end of offline timestep [H ~> m or kg m-2]accumulated_time: Length of time accumulated in the current offline interval [s][out] dt_offline: Timestep used for offline tracers [s][out] dt_offline_vertical: Timestep used for calls to tracer vertical physics [s][out] skip_diffusion: Skips horizontal diffusion of tracers
-
subroutine, public mom_offline_main::insert_offline_main(CS CS, ALE_CSp ALE_CSp, diabatic_CSp diabatic_CSp, diag diag, OBC OBC, tracer_adv_CSp tracer_adv_CSp, tracer_flow_CSp tracer_flow_CSp, tracer_Reg tracer_Reg, tv tv, G G, GV GV, x_before_y x_before_y, debug debug) Inserts (assigns values to) members of the offline main control structure. All arguments are optional except for the CS itself.
- Parameters
[inout] cs: Offline control structure[in] ale_csp: A pointer to the ALE control structure[in] diabatic_csp: A pointer to the diabatic control structure[in] diag: A pointer to the structure that regulates diagnostic output[in] obc: A pointer to the open boundary condition control structure[in] tracer_adv_csp: A pointer to the tracer advection control structure[in] tracer_flow_csp: A pointer to the tracer flow control control structure[in] tracer_reg: A pointer to the tracer registry[in] tv: A structure pointing to various thermodynamic variables[in] g: ocean grid structure[in] gv: ocean vertical grid structure[in] x_before_y: Indicates which horizontal direction is advected first[in] debug: If true, write verbose debugging messages
-
subroutine, public mom_offline_main::offline_transport_init(param_file param_file, CS CS, diabatic_CSp diabatic_CSp, G G, GV GV) Initializes the control structure for offline transport and reads in some of the.
- Parameters
[in] param_file: A structure to parse for run-time parameterscs: Offline control structure[in] diabatic_csp: The diabatic control structure[in] g: ocean grid structure[in] gv: ocean vertical grid structure
-
subroutine
read_all_input(CS CS)¶ Coordinates the allocation and reading in all time levels of uh, vh, hend, temp, and salt from files. Used when read_all_ts_uvh.
- Parameters
[inout] cs: Control structure for offline module
-
subroutine, public mom_offline_main::offline_transport_end(CS CS) Deallocates (if necessary) arrays within the offline control structure.
- Parameters
cs: Control structure for offline module
-
-
namespace
mom_opacity¶ Routines used to calculate the opacity of the ocean.
opacity_from_chl: In this routine, the Morel (modified) or Manizza (modified) schemes use the “blue” band in the paramterizations to determine the e-folding depth of the incoming shortwave attenuation. The red portion is lumped into the net heating at the surface.
Morel, A., 1988: Optical modeling of the upper ocean in relation to its biogenous matter content (case-i waters)., J. Geo. Res., 93, 10,749-10,768.
Manizza, M., C. LeQuere, A. J. Watson, and E. T. Buitenhuis, 2005: Bio-optical feedbacks among phytoplankton, upper ocean physics and sea-ice in a global model, Geophys. Res. Let., 32, L05603, doi:10.1029/2004GL020778.
Unnamed Group
-
integer, parameter mom_opacity::no_scheme = 0 Coded integers to specify the opacity scheme.
-
integer, parameter mom_opacity::manizza_05 = 1 Coded integers to specify the opacity scheme.
-
integer, parameter mom_opacity::morel_88 = 2 Coded integers to specify the opacity scheme.
-
integer, parameter mom_opacity::single_exp = 3 Coded integers to specify the opacity scheme.
-
integer, parameter mom_opacity::double_exp = 4 Coded integers to specify the opacity scheme.
-
character*(10), parameter mom_opacity::manizza_05_string = "MANIZZA_05" String to specify the opacity scheme.
-
character*(10), parameter mom_opacity::morel_88_string = "MOREL_88" String to specify the opacity scheme.
-
character*(10), parameter mom_opacity::single_exp_string = "SINGLE_EXP" String to specify the opacity scheme.
-
character*(10), parameter mom_opacity::double_exp_string = "DOUBLE_EXP" String to specify the opacity scheme.
-
real, parameter mom_opacity::op_diag_len = 1e-10 Lengthscale L used to remap opacity from op to 1/L * tanh(op * L)
-
subroutine, public mom_opacity::set_opacity(optics optics, sw_total sw_total, sw_vis_dir sw_vis_dir, sw_vis_dif sw_vis_dif, sw_nir_dir sw_nir_dir, sw_nir_dif sw_nir_dif, G G, GV GV, CS CS, chl_2d chl_2d, chl_3d chl_3d) This sets the opacity of sea water based based on one of several different schemes.
- Parameters
[inout] optics: An optics structure that has values set based on the opacities.sw_total: Total shortwave flux into the ocean [W m-2]sw_vis_dir: Visible, direct shortwave into the ocean [W m-2]sw_vis_dif: Visible, diffuse shortwave into the ocean [W m-2]sw_nir_dir: Near-IR, direct shortwave into the ocean [W m-2]sw_nir_dif: Near-IR, diffuse shortwave into the ocean [W m-2][in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.cs: The control structure earlier set up by opacity_init.[in] chl_2d: Vertically uniform chlorophyll-A concentractions[mg m-3][in] chl_3d: The chlorophyll-A concentractions of each layer [mg m-3]
-
subroutine
opacity_from_chl(optics optics, sw_total sw_total, sw_vis_dir sw_vis_dir, sw_vis_dif sw_vis_dif, sw_nir_dir sw_nir_dir, sw_nir_dif sw_nir_dif, G G, GV GV, CS CS, chl_2d chl_2d, chl_3d chl_3d)¶ This sets the “blue” band opacity based on chloophyll A concencentrations The red portion is lumped into the net heating at the surface.
- Parameters
[inout] optics: An optics structure that has values set based on the opacities.sw_total: Total shortwave flux into the ocean [W m-2]sw_vis_dir: Visible, direct shortwave into the ocean [W m-2]sw_vis_dif: Visible, diffuse shortwave into the ocean [W m-2]sw_nir_dir: Near-IR, direct shortwave into the ocean [W m-2]sw_nir_dif: Near-IR, diffuse shortwave into the ocean [W m-2][in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.cs: The control structure.[in] chl_2d: Vertically uniform chlorophyll-A concentractions [mg m-3][in] chl_3d: A 3-d field of chlorophyll-A concentractions [mg m-3]
-
real function, public mom_opacity::opacity_morel(chl_data chl_data) This sets the blue-wavelength opacity according to the scheme proposed by Morel and Antoine (1994).
- Return
The returned opacity [m-1]
- Parameters
[in] chl_data: The chlorophyll-A concentration in mg m-3.
-
real function mom_opacity::sw_pen_frac_morel(chl_data chl_data) This sets the penetrating shortwave fraction according to the scheme proposed by Morel and Antoine (1994).
- Return
The returned penetrating shortwave fraction [nondim]
- Parameters
[in] chl_data: The chlorophyll-A concentration in mg m-3.
-
real function, public mom_opacity::opacity_manizza(chl_data chl_data) This sets the blue-wavelength opacity according to the scheme proposed by Manizza, M. et al, 2005.
- Return
The returned opacity [m-1]
- Parameters
[in] chl_data: The chlorophyll-A concentration in mg m-3.
-
subroutine, public mom_opacity::extract_optics_slice(optics optics, j j, G G, GV GV, opacity opacity, opacity_scale opacity_scale, penSW_top penSW_top, penSW_scale penSW_scale) This subroutine returns a 2-d slice at constant j of fields from an optics_type, with the potential for rescaling these fields.
- Parameters
[in] optics: An optics structure that has values of opacities and shortwave fluxes.[in] j: j-index to extract[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[out] opacity: The opacity in each band, i-point, and layer[in] opacity_scale: A factor by which to rescale the opacity.[out] pensw_top: The shortwave radiation [W m-2] at the surface[in] pensw_scale: A factor by which to rescale the shortwave flux.
-
subroutine, public mom_opacity::extract_optics_fields(optics optics, nbands nbands) Set arguments to fields from the optics type.
- Parameters
[in] optics: An optics structure that has values of opacities and shortwave fluxes.[out] nbands: The number of penetrating bands of SW radiation
-
integer function, public mom_opacity::optics_nbands(optics optics) Return the number of bands of penetrating shortwave radiation.
- Return
The number of penetrating bands of SW radiation
- Parameters
[in] optics: An optics structure that has values of opacities and shortwave fluxes.
-
subroutine, public mom_opacity::absorbremainingsw(G G, GV GV, US US, h h, opacity_band opacity_band, nsw nsw, optics optics, j j, dt dt, H_limit_fluxes H_limit_fluxes, adjustAbsorptionProfile adjustAbsorptionProfile, absorbAllSW absorbAllSW, T T, Pen_SW_bnd Pen_SW_bnd, eps eps, ksort ksort, htot htot, Ttot Ttot, TKE TKE, dSV_dT dSV_dT) Apply shortwave heating below the boundary layer (when running with the bulk mixed layer inhereted from GOLD) or throughout the water column.
In addition, it causes all of the remaining SW radiation to be absorbed, provided that the total water column thickness is greater than H_limit_fluxes. For thinner water columns, the heating is scaled down proportionately, the assumption being that the remaining heating (which is left in Pen_SW) should go into an (absent for now) ocean bottom sediment layer.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] nsw: Number of bands of penetrating shortwave radiation.[in] h: Layer thicknesses [H ~> m or kg m-2].[in] opacity_band: Opacity in each band of penetrating shortwave radiation [H-1 ~> m-1 or m2 kg-1]. The indicies are band, i, k.[in] optics: An optics structure that has values of opacities and shortwave fluxes.[in] j: j-index to work on.[in] dt: Time step [T ~> s].[in] h_limit_fluxes: If the total ocean depth is less than this, they are scaled away to avoid numerical instabilities [H ~> m or kg m-2]. This would not be necessary if a finite heat capacity mud-layer were added.[in] adjustabsorptionprofile: If true, apply heating above the layers in which it should have occurred to get the correct mean depth (and potential energy change) of the shortwave that should be absorbed by each layer.[in] absorballsw: If true, apply heating above the layers in which it should have occurred to get the correct mean depth (and potential energy change) of the shortwave that should be absorbed by each layer.[inout] t: Layer potential/conservative temperatures [degC][inout] pen_sw_bnd: Penetrating shortwave heating in each band that hits the bottom and will will be redistributed through the water column [degC H ~> degC m or degC kg m-2], size nsw x G isd: G ied.[in] eps: Small thickness that must remain in each layer, and which will not be subject to heating [H ~> m or kg m-2][in] ksort: Density-sorted k-indicies.[in] htot: Total mixed layer thickness [H ~> m or kg m-2].[inout] ttot: Depth integrated mixed layer temperature [degC H ~> degC m or degC kg m-2][in] dsv_dt: The partial derivative of specific volume with temperature [m3 kg-1 degC-1].[inout] tke: The TKE sink from mixing the heating throughout a layer [kg m-3 Z3 T-2 ~> J m-2].
-
subroutine, public mom_opacity::sumswoverbands(G G, GV GV, US US, h h, nsw nsw, optics optics, j j, dt dt, H_limit_fluxes H_limit_fluxes, absorbAllSW absorbAllSW, iPen_SW_bnd iPen_SW_bnd, netPen netPen) This subroutine calculates the total shortwave heat flux integrated over bands as a function of depth. This routine is only called for computing buoyancy fluxes for use in KPP. This routine does not updat e the state.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] h: Layer thicknesses [H ~> m or kg m-2].[in] nsw: The number of bands of penetrating shortwave radiation, perhaps from optics_nbands(optics),[in] optics: An optics structure that has values set based on the opacities.[in] j: j-index to work on.[in] dt: Time step [T ~> s].[in] h_limit_fluxes: the total depth at which the surface fluxes start to be limited to avoid excessive heating of a thin ocean [H ~> m or kg m-2][in] absorballsw: If true, ensure that all shortwave radiation is absorbed in the ocean water column.[in] ipen_sw_bnd: The incident penetrating shortwave heating in each band that hits the bottom and will be redistributed through the water column [degC H ~> degC m or degC kg m-2]; size nsw x G isd: G ied.[inout] netpen: Net penetrating shortwave heat flux at each
-
subroutine, public mom_opacity::opacity_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS, optics optics) This routine initalizes the opacity module, including an optics_type.
- Parameters
[in] time: The current model time.[in] g: The ocean’s grid structure.[in] gv: model vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters.[inout] diag: A structure that is used to regulate diagnostic output.cs: A pointer that is set to point to the control structure for this module.optics: An optics structure that has parameters set and arrays allocated here.
-
subroutine, public mom_opacity::opacity_end(CS CS, optics optics) - Parameters
cs: An opacity control structure that should be deallocated.optics: An optics type structure that should be deallocated.
-
-
namespace
mom_open_boundary¶ Controls where open boundary conditions are applied.
This module implements some aspects of internal open boundary conditions in MOM.
A small fragment of the grid is shown below:
j+1 x ^ x ^ x At x: q, CoriolisBu j+1 > o > o > At ^: v, tauy j x ^ x ^ x At >: u, taux j > o > o > At o: h, bathyT, buoy, tr, T, S, Rml, ustar j-1 x ^ x ^ x i-1 i i+1 At x & ^: i i+1 At > & o:
The boundaries always run through q grid points (x).
Functions
-
subroutine, public mom_open_boundary::open_boundary_config(G G, US US, param_file param_file, OBC OBC) Enables OBC module and reads configuration parameters This routine is called from MOM_initialize_fixed which occurs before the initialization of the vertical coordinate and ALE_init. Therefore segment data are not fully initialized here. The remainder of the segment data are initialized in a later call to update_open_boundary_data.
- Parameters
[inout] g: Ocean grid structure[in] us: A dimensional unit scaling type[in] param_file: Parameter file handleobc: Open boundary control structure
-
subroutine
initialize_segment_data(G G, OBC OBC, PF PF)¶ Allocate space for reading OBC data from files. It sets up the required vertical remapping. In the process, it does funky stuff with the MPI processes.
- Parameters
[in] g: Ocean grid structure[inout] obc: Open boundary control structure[in] pf: Parameter file handle
-
subroutine
setup_segment_indices(G G, seg seg, Is_obc Is_obc, Ie_obc Ie_obc, Js_obc Js_obc, Je_obc Je_obc)¶ Define indices for segment and store in hor_index_type using global segment bounds corresponding to q-points.
- Parameters
[in] g: grid type[inout] seg: Open boundary segment[in] is_obc: Q-point global i-index of start of segment[in] ie_obc: Q-point global i-index of end of segment[in] js_obc: Q-point global j-index of start of segment[in] je_obc: Q-point global j-index of end of segment
-
subroutine
setup_u_point_obc(OBC OBC, G G, segment_str segment_str, l_seg l_seg, PF PF, reentrant_y reentrant_y)¶ Parse an OBC_SEGMENT_%%% string starting with “I=” and configure placement and type of OBC accordingly.
- Parameters
obc: Open boundary control structure[in] g: Ocean grid structure[in] segment_str: A string in form of “I=%,J=%:%,string”[in] l_seg: which segment is this?[in] pf: Parameter file handle[in] reentrant_y: is the domain reentrant in y?
-
subroutine
setup_v_point_obc(OBC OBC, G G, segment_str segment_str, l_seg l_seg, PF PF, reentrant_x reentrant_x)¶ Parse an OBC_SEGMENT_%%% string starting with “J=” and configure placement and type of OBC accordingly.
- Parameters
obc: Open boundary control structure[in] g: Ocean grid structure[in] segment_str: A string in form of “J=%,I=%:%,string”[in] l_seg: which segment is this?[in] pf: Parameter file handle[in] reentrant_x: is the domain reentrant in x?
-
subroutine
parse_segment_str(ni_global ni_global, nj_global nj_global, segment_str segment_str, l l, m m, n n, action_str action_str, reentrant reentrant)¶ Parse an OBC_SEGMENT_%%% string.
- Parameters
[in] ni_global: Number of h-points in zonal direction[in] nj_global: Number of h-points in meridional direction[in] segment_str: A string in form of “I=l,J=m:n,string” or “J=l,I=m,n,string”[out] l: The value of I=l, if segment_str begins with I=l, or the value of J=l[out] m: The value of J=m, if segment_str begins with I=, or the value of I=m[out] n: The value of J=n, if segment_str begins with I=, or the value of I=n[out] action_str: The “string” part of segment_str[in] reentrant: is domain reentrant in relevant direction?
-
subroutine
parse_segment_data_str(segment_str segment_str, var var, value value, filenam filenam, fieldnam fieldnam, fields fields, num_fields num_fields, debug debug)¶ Parse an OBC_SEGMENT_%%_DATA string.
- Parameters
[in] segment_str: A string in form of “VAR1=file:foo1.nc(varnam1),VAR2=file:foo2.nc(varnam2),…”[in] var: The name of the variable for which parameters are needed[out] filenam: The name of the input file if using “file” method[out] fieldnam: The name of the variable in the input file if using “file” method[out] value: A constant value if using the “value” method[out] fields: List of fieldnames for each segment[out] num_fields: The number of fields in the segment data[in] debug: If present and true, write verbose debugging messages
-
subroutine
parse_segment_param_real(segment_str segment_str, var var, param_value param_value, debug debug)¶ Parse an OBC_SEGMENT_%%_PARAMS string.
- Parameters
[in] segment_str: A string in form of “VAR1=file:foo1.nc(varnam1),VAR2=file:foo2.nc(varnam2),…”[in] var: The name of the variable for which parameters are needed[out] param_value: The value of the parameter[in] debug: If present and true, write verbose debugging messages
-
subroutine, public mom_open_boundary::open_boundary_init(G G, param_file param_file, OBC OBC) Initialize open boundary control structure.
- Parameters
[in] g: Ocean grid structure[in] param_file: Parameter file handleobc: Open boundary control structure
-
logical function, public mom_open_boundary::open_boundary_query(OBC OBC, apply_open_OBC apply_open_OBC, apply_specified_OBC apply_specified_OBC, apply_Flather_OBC apply_Flather_OBC, apply_nudged_OBC apply_nudged_OBC, needs_ext_seg_data needs_ext_seg_data) - Parameters
obc: Open boundary control structure[in] apply_open_obc: Returns True if open_*_BCs_exist_globally is true[in] apply_specified_obc: Returns True if specified_*_BCs_exist_globally is true[in] apply_flather_obc: Returns True if Flather_*_BCs_exist_globally is true[in] apply_nudged_obc: Returns True if nudged_*_BCs_exist_globally is true[in] needs_ext_seg_data: Returns True if external segment data needed
-
subroutine
open_boundary_dealloc(OBC OBC)¶ Deallocate open boundary data.
- Parameters
obc: Open boundary control structure
-
subroutine, public mom_open_boundary::open_boundary_end(OBC OBC) Close open boundary data.
- Parameters
obc: Open boundary control structure
-
subroutine, public mom_open_boundary::open_boundary_impose_normal_slope(OBC OBC, G G, depth depth) Sets the slope of bathymetry normal to an open bounndary to zero.
- Parameters
obc: Open boundary control structure[in] g: Ocean grid structure[inout] depth: Bathymetry at h-points
-
subroutine, public mom_open_boundary::open_boundary_impose_land_mask(OBC OBC, G G, areaCu areaCu, areaCv areaCv) Reconcile masks and open boundaries, deallocate OBC on PEs where it is not needed. Also adjust u- and v-point cell area on specified open boundaries and mask all points outside open boundaries.
- Parameters
obc: Open boundary control structure[inout] g: Ocean grid structure[inout] areacu: Area of a u-cell [m2][inout] areacv: Area of a u-cell [m2]
-
subroutine, public mom_open_boundary::radiation_open_bdry_conds(OBC OBC, u_new u_new, u_old u_old, v_new v_new, v_old v_old, G G, dt dt) Apply radiation conditions to 3D u,v at open boundaries.
- Parameters
[inout] g: Ocean grid structureobc: Open boundary control structure[inout] u_new: On exit, new u values on open boundaries On entry, the old time-level v but including barotropic accelerations.[in] u_old: Original unadjusted u[inout] v_new: On exit, new v values on open boundaries. On entry, the old time-level v but including barotropic accelerations.[in] v_old: Original unadjusted v[in] dt: Appropriate timestep
-
subroutine, public mom_open_boundary::open_boundary_apply_normal_flow(OBC OBC, G G, u u, v v) Applies OBC values stored in segments to 3d u,v fields.
- Parameters
obc: Open boundary control structure[inout] g: Ocean grid structure[inout] u: u field to update on open boundaries[inout] v: v field to update on open boundaries
-
subroutine, public mom_open_boundary::open_boundary_zero_normal_flow(OBC OBC, G G, u u, v v) Applies zero values to 3d u,v fields on OBC segments.
- Parameters
obc: Open boundary control structure[inout] g: Ocean grid structure[inout] u: u field to update on open boundaries[inout] v: v field to update on open boundaries
-
subroutine
gradient_at_q_points(G G, segment segment, uvel uvel, vvel vvel)¶ Calculate the tangential gradient of the normal flow at the boundary q-points.
- Parameters
[in] g: Ocean grid structuresegment: OBC segment structure[in] uvel: zonal velocity[in] vvel: meridional velocity
-
subroutine, public mom_open_boundary::set_tracer_data(OBC OBC, tv tv, h h, G G, PF PF, tracer_Reg tracer_Reg) Sets the initial values of the tracer open boundary conditions. Redoing this elsewhere.
- Parameters
[inout] g: Ocean grid structureobc: Open boundary structure[inout] tv: Thermodynamics structure[inout] h: Thickness[in] pf: Parameter file handletracer_reg: Tracer registry
-
integer function mom_open_boundary::lookup_seg_field(OBC_seg OBC_seg, field field) Needs documentation.
- Parameters
obc_seg: OBC segment[in] field: The field name
-
subroutine
allocate_obc_segment_data(OBC OBC, segment segment)¶ Allocate segment data fields.
- Parameters
obc: Open boundary structure[inout] segment: Open boundary segment
-
subroutine
deallocate_obc_segment_data(OBC OBC, segment segment)¶ Deallocate segment data fields.
- Parameters
obc: Open boundary structure[inout] segment: Open boundary segment
-
subroutine, public mom_open_boundary::open_boundary_test_extern_uv(G G, OBC OBC, u u, v v) Set tangential velocities outside of open boundaries to silly values (used for checking the interior state is independent of values outside of the domain).
- Parameters
[in] g: Ocean grid structureobc: Open boundary structure[inout] u: Zonal velocity [m s-1][inout] v: Meridional velocity [m s-1]
-
subroutine, public mom_open_boundary::open_boundary_test_extern_h(G G, OBC OBC, h h) Set thicknesses outside of open boundaries to silly values (used for checking the interior state is independent of values outside of the domain).
- Parameters
[in] g: Ocean grid structureobc: Open boundary structure[inout] h: Layer thickness [H ~> m or kg m-2]
-
subroutine, public mom_open_boundary::update_obc_segment_data(G G, GV GV, US US, OBC OBC, tv tv, h h, Time Time) Update the OBC values on the segments.
- Parameters
[in] g: Ocean grid structure[in] gv: Ocean vertical grid structure[in] us: A dimensional unit scaling typeobc: Open boundary structure[in] tv: Thermodynamics structure[inout] h: Thickness [m][in] time: Model time
-
subroutine, public mom_open_boundary::register_obc(name name, param_file param_file, Reg Reg) register open boundary objects for boundary updates.
- Parameters
[in] name: OBC name used for error messages[in] param_file: file to parse for model parameter valuesreg: pointer to the tracer registry
-
subroutine, public mom_open_boundary::obc_registry_init(param_file param_file, Reg Reg) This routine include declares and sets the variable “version”.
- Parameters
[in] param_file: open file to parse for model parametersreg: pointer to OBC registry
-
logical function, public mom_open_boundary::register_file_obc(param_file param_file, CS CS, OBC_Reg OBC_Reg) Add file to OBC registry.
- Parameters
[in] param_file: parameter file.cs: file control structure.obc_reg: OBC registry.
-
subroutine, public mom_open_boundary::file_obc_end(CS CS) Clean up the file OBC from registry.
- Parameters
cs: OBC file control structure.
-
subroutine, public mom_open_boundary::segment_tracer_registry_init(param_file param_file, segment segment) Initialize the segment tracer registry.
- Parameters
[in] param_file: open file to parse for model parameters[inout] segment: the segment
-
subroutine, public mom_open_boundary::register_segment_tracer(tr_ptr tr_ptr, param_file param_file, GV GV, segment segment, OBC_scalar OBC_scalar, OBC_array OBC_array) - Parameters
[in] gv: ocean vertical grid structuretr_ptr: A target that can be used to set a pointer to the stored value of tr. This target must be an enduring part of the control structure, because the tracer registry will use this memory, but it also means that any updates to this structure in the calling module will be available subsequently to the tracer registry.[in] param_file: file to parse for model parameter values[inout] segment: current segment data structure[in] obc_scalar: If present, use scalar value for segment tracer inflow concentration.[in] obc_array: If true, use array values for segment tracer inflow concentration.
-
subroutine, public mom_open_boundary::segment_tracer_registry_end(Reg Reg) Clean up the segment tracer registry.
- Parameters
reg: pointer to tracer registry
-
subroutine, public mom_open_boundary::register_temp_salt_segments(GV GV, OBC OBC, tr_Reg tr_Reg, param_file param_file) - Parameters
[in] gv: ocean vertical grid structureobc: Open boundary structuretr_reg: Tracer registry[in] param_file: file to parse for model parameter values
-
subroutine, public mom_open_boundary::fill_temp_salt_segments(G G, OBC OBC, tv tv) - Parameters
[inout] g: Ocean grid structureobc: Open boundary structure[inout] tv: Thermodynamics structure
-
subroutine
mask_outside_obcs(G G, US US, param_file param_file, OBC OBC)¶ Find the region outside of all open boundary segments and make sure it is set to land mask. Gonna need to know global land mask as well to get it right…
- Parameters
[inout] g: Ocean grid structure[in] param_file: Parameter file handleobc: Open boundary structure[in] us: A dimensional unit scaling type
-
subroutine
flood_fill(G G, color color, cin cin, cout cout, cland cland)¶ flood the cin, cout values
- Parameters
[inout] g: Ocean grid structure[inout] color: For sorting inside from outside[in] cin: color for inside the domain[in] cout: color for outside the domain[in] cland: color for inside the land mask
-
subroutine
flood_fill2(G G, color color, cin cin, cout cout, cland cland)¶ flood the cin, cout values
- Parameters
[inout] g: Ocean grid structure[inout] color: For sorting inside from outside[in] cin: color for inside the domain[in] cout: color for outside the domain[in] cland: color for inside the land mask
-
subroutine, public mom_open_boundary::open_boundary_register_restarts(HI HI, GV GV, OBC_CS OBC_CS, restart_CSp restart_CSp) Register OBC segment data for restarts.
- Parameters
[in] hi: Horizontal indicesgv: Container for vertical grid informationobc_cs: OBC data structure, data intent(inout)restart_csp: Restart structure, data intent(inout)
Variables
-
integer, parameter, public mom_open_boundary::obc_none = 0 Indicates the use of no open boundary.
-
integer, parameter, public mom_open_boundary::obc_simple = 1 Indicates the use of a simple inflow open boundary.
-
integer, parameter, public mom_open_boundary::obc_wall = 2 Indicates the use of a closed sall.
-
integer, parameter, public mom_open_boundary::obc_flather = 3 Indicates the use of a Flather open boundary.
-
integer, parameter, public mom_open_boundary::obc_radiation = 4 Indicates the use of a radiation open boundary.
-
integer, parameter, public mom_open_boundary::obc_direction_n = 100 Indicates the boundary is an effective northern boundary.
-
integer, parameter, public mom_open_boundary::obc_direction_s = 200 Indicates the boundary is an effective southern boundary.
-
integer, parameter, public mom_open_boundary::obc_direction_e = 300 Indicates the boundary is an effective eastern boundary.
-
integer, parameter, public mom_open_boundary::obc_direction_w = 400 Indicates the boundary is an effective western boundary.
-
integer, parameter mom_open_boundary::max_obc_fields = 100 Maximum number of data fields needed for OBC segments.
-
integer
id_clock_pass¶ A CPU time clock.
-
character(len=40) mom_open_boundary::mdl = "MOM_open_boundary" This module’s name.
-
-
namespace
mom_pointaccel¶ Debug accelerations at a given point.
The two subroutines in this file write out all of the terms in the u- or v-momentum balance at a given point. Usually these subroutines are called after the velocities exceed some threshold, in order to determine which term is culpable. often this is done for debugging purposes.
Functions
-
subroutine, public mom_pointaccel::write_u_accel(I I, j j, um um, hin hin, ADp ADp, CDp CDp, dt dt, G G, GV GV, US US, CS CS, vel_rpt vel_rpt, str str, a a, hv hv) This subroutine writes to an output file all of the accelerations that have been applied to a column of zonal velocities over the previous timestep. This subroutine is called from vertvisc.
- Parameters
[in] i: The zonal index of the column to be documented.[in] j: The meridional index of the column to be documented.[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] um: The new zonal velocity [m s-1].[in] hin: The layer thickness [H ~> m or kg m-2].[in] adp: A structure pointing to the various accelerations in the momentum equations.[in] cdp: A structure with pointers to various terms in the continuity equations.[in] dt: The ocean dynamics time step [s].cs: The control structure returned by a previous call to PointAccel_init.[in] vel_rpt: The velocity magnitude that triggers a report [m s-1].[in] str: The surface wind stress integrated over a time step divided by the Boussinesq density [m2 s-1].[in] a: The layer coupling coefficients from vertvisc [Z s-1 ~> m s-1].[in] hv: The layer thicknesses at velocity grid points,
-
subroutine, public mom_pointaccel::write_v_accel(i i, J J, vm vm, hin hin, ADp ADp, CDp CDp, dt dt, G G, GV GV, US US, CS CS, vel_rpt vel_rpt, str str, a a, hv hv) This subroutine writes to an output file all of the accelerations that have been applied to a column of meridional velocities over the previous timestep. This subroutine is called from vertvisc.
- Parameters
[in] i: The zonal index of the column to be documented.[in] j: The meridional index of the column to be documented.[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] vm: The new meridional velocity [m s-1].[in] hin: The layer thickness [H ~> m or kg m-2].[in] adp: A structure pointing to the various accelerations in the momentum equations.[in] cdp: A structure with pointers to various terms in the continuity equations.[in] dt: The ocean dynamics time step [s].cs: The control structure returned by a previous call to PointAccel_init.[in] vel_rpt: The velocity magnitude that triggers a report [m s-1].[in] str: The surface wind stress integrated over a time step divided by the Boussinesq density [m2 s-1].[in] a: The layer coupling coefficients from vertvisc [Z s-1 ~> m s-1].[in] hv: The layer thicknesses at velocity grid points,
-
subroutine, public mom_pointaccel::pointaccel_init(MIS MIS, Time Time, G G, param_file param_file, diag diag, dirs dirs, CS CS) This subroutine initializes the parameters regulating how truncations are logged.
- Parameters
[in] mis: For “MOM Internal State” a set of pointers[in] time: The current model time.[in] g: The ocean’s grid structure.[in] param_file: A structure to parse for run-time parameters.[inout] diag: A structure that is used to regulate diagnostic output.[in] dirs: A structure containing several relevant directory paths.cs: A pointer that is set to point to the control structure for this module.
-
-
namespace
mom_pressureforce¶ A thin wrapper for Boussinesq/non-Boussinesq forms of the pressure force calculation.
This thin module provides a branch to two forms of the horizontal accelerations due to pressure gradients. The two options currently available are a Montgomery potential form (used in traditional isopycnal layer models), and the analytic finite volume form.
Functions
-
subroutine, public mom_pressureforce::pressureforce(h h, tv tv, PFu PFu, PFv PFv, G G, GV GV, US US, CS CS, ALE_CSp ALE_CSp, p_atm p_atm, pbce pbce, eta eta) A thin layer between the model and the Boussinesq and non-Boussinesq pressure force routines.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[in] h: Layer thicknesses [H ~> m or kg m-2][in] tv: A structure pointing to various thermodynamic variables[out] pfu: Zonal pressure force acceleration [m s-2][out] pfv: Meridional pressure force acceleration [m s-2]cs: Pressure force control structureale_csp: ALE control structurep_atm: The pressure at the ice-ocean or[out] pbce: The baroclinic pressure anomaly in each layer[out] eta: The bottom mass used to calculate PFu and PFv,
-
subroutine, public mom_pressureforce::pressureforce_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS, tides_CSp tides_CSp) Initialize the pressure force control structure.
- Parameters
[in] time: Current model time[in] g: Ocean grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: Parameter file handles[inout] diag: Diagnostics control structurecs: Pressure force control structuretides_csp: Tide control structure
-
subroutine, public mom_pressureforce::pressureforce_end(CS CS) Deallocate the pressure force control structure.
- Parameters
cs: Pressure force control structure
-
-
namespace
mom_pressureforce_afv¶ Analytically integrated finite volume pressure gradient.
Provides the Boussinesq and non-Boussinesq forms of horizontal accelerations due to pressure gradients using a 2nd-order analytically vertically integrated finite volume form, as described by Adcroft et al., 2008.
This form eliminates the thermobaric instabilities that had been a problem with previous forms of the pressure gradient force calculation, as described by Hallberg, 2005.
Adcroft, A., R. Hallberg, and M. Harrison, 2008: A finite volume discretization of the pressure gradient force using analytic integration. Ocean Modelling, 22, 106-113. http://doi.org/10.1016/j.ocemod.2008.02.001
Hallberg, 2005: A thermobaric instability of Lagrangian vertical coordinate ocean models. Ocean Modelling, 8, 279-300. http://dx.doi.org/10.1016/j.ocemod.2004.01.001
Functions
-
subroutine, public mom_pressureforce_afv::pressureforce_afv(h h, tv tv, PFu PFu, PFv PFv, G G, GV GV, US US, CS CS, ALE_CSp ALE_CSp, p_atm p_atm, pbce pbce, eta eta) Thin interface between the model and the Boussinesq and non-Boussinesq pressure force routines.
- Parameters
[in] g: Ocean grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[in] h: Layer thickness [H ~> m or kg m-2][inout] tv: Thermodynamic variables[out] pfu: Zonal acceleration [m s-2][out] pfv: Meridional acceleration [m s-2]cs: Finite volume PGF control structureale_csp: ALE control structurep_atm: The pressure at the ice-ocean or atmosphere-ocean interface [Pa].[out] pbce: The baroclinic pressure anomaly in each layer due to eta anomalies [m2 s-2 H-1 ~> m s-2 or m4 s-2 kg-1].[out] eta: The bottom mass used to calculate PFu and PFv [H ~> m or kg m-2], with any tidal contributions or compressibility compensation.
-
subroutine, public mom_pressureforce_afv::pressureforce_afv_nonbouss(h h, tv tv, PFu PFu, PFv PFv, G G, GV GV, US US, CS CS, ALE_CSp ALE_CSp, p_atm p_atm, pbce pbce, eta eta) Non-Boussinesq analytically-integrated finite volume form of pressure gradient.
Determines the acceleration due to hydrostatic pressure forces, using the analytic finite volume form of the Pressure gradient, and does not make the Boussinesq approximation.
To work, the following fields must be set outside of the usual (is:ie,js:je) range before this subroutine is called: h(isB:ie+1,jsB:je+1), T(isB:ie+1,jsB:je+1), and S(isB:ie+1,jsB:je+1).
- Parameters
[in] g: Ocean grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[in] h: Layer thickness [H ~> kg/m2][in] tv: Thermodynamic variables[out] pfu: Zonal acceleration [m s-2][out] pfv: Meridional acceleration [m s-2]cs: Finite volume PGF control structureale_csp: ALE control structurep_atm: The pressure at the ice-ocean or atmosphere-ocean interface [Pa].[out] pbce: The baroclinic pressure anomaly in each layer due to eta anomalies [m2 s-2 H-1 ~> m s-2 or m4 s-2 kg-1].[out] eta: The bottom mass used to calculate PFu and PFv [H ~> m or kg m-2], with any tidal contributions or compressibility compensation.
-
subroutine, public mom_pressureforce_afv::pressureforce_afv_bouss(h h, tv tv, PFu PFu, PFv PFv, G G, GV GV, US US, CS CS, ALE_CSp ALE_CSp, p_atm p_atm, pbce pbce, eta eta) Boussinesq analytically-integrated finite volume form of pressure gradient.
Determines the acceleration due to hydrostatic pressure forces, using the finite volume form of the terms and analytic integrals in depth.
To work, the following fields must be set outside of the usual (is:ie,js:je) range before this subroutine is called: h(isB:ie+1,jsB:je+1), T(isB:ie+1,jsB:je+1), and S(isB:ie+1,jsB:je+1).
- Parameters
[in] g: Ocean grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[in] h: Layer thickness [H ~> m][in] tv: Thermodynamic variables[out] pfu: Zonal acceleration [m s-2][out] pfv: Meridional acceleration [m s-2]cs: Finite volume PGF control structureale_csp: ALE control structurep_atm: The pressure at the ice-ocean or atmosphere-ocean interface [Pa].[out] pbce: The baroclinic pressure anomaly in each layer due to eta anomalies [m2 s-2 H-1 ~> m s-2 or m4 s-2 kg-1].[out] eta: The bottom mass used to calculate PFu and PFv [H ~> m or kg m-2], with any tidal contributions or compressibility compensation.
-
subroutine, public mom_pressureforce_afv::pressureforce_afv_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS, tides_CSp tides_CSp) Initializes the finite volume pressure gradient control structure.
- Parameters
[in] time: Current model time[in] g: Ocean grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: Parameter file handles[inout] diag: Diagnostics control structurecs: Finite volume PGF control structuretides_csp: Tides control structure
-
subroutine, public mom_pressureforce_afv::pressureforce_afv_end(CS CS) Deallocates the finite volume pressure gradient control structure.
- Parameters
cs: Finite volume pressure control structure that will be deallocated in this subroutine.
-
-
namespace
mom_pressureforce_blk_afv¶ Analytically integrated finite volume pressure gradient.
Functions
-
subroutine, public mom_pressureforce_blk_afv::pressureforce_blk_afv(h h, tv tv, PFu PFu, PFv PFv, G G, GV GV, US US, CS CS, ALE_CSp ALE_CSp, p_atm p_atm, pbce pbce, eta eta) Thin interface between the model and the Boussinesq and non-Boussinesq pressure force routines.
- Parameters
[in] g: Ocean grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[in] h: Layer thickness [H ~> m or kg m-2][inout] tv: Thermodynamic variables[out] pfu: Zonal acceleration [m s-2][out] pfv: Meridional acceleration [m s-2]cs: Finite volume PGF control structureale_csp: ALE control structurep_atm: The pressure at the ice-ocean or atmosphere-ocean interface [Pa].[out] pbce: The baroclinic pressure anomaly in each layer due to eta anomalies [m2 s-2 H-1 ~> m s-2 or m4 s-2 kg-1].[out] eta: The bottom mass used to calculate PFu and PFv [H ~> m or kg m-2], with any tidal contributions or compressibility compensation.
-
subroutine, public mom_pressureforce_blk_afv::pressureforce_blk_afv_nonbouss(h h, tv tv, PFu PFu, PFv PFv, G G, GV GV, US US, CS CS, p_atm p_atm, pbce pbce, eta eta) Non-Boussinesq analytically-integrated finite volume form of pressure gradient.
Determines the acceleration due to hydrostatic pressure forces, using the analytic finite volume form of the Pressure gradient, and does not make the Boussinesq approximation. This version uses code-blocking for threads.
To work, the following fields must be set outside of the usual (is:ie,js:je) range before this subroutine is called: h(isB:ie+1,jsB:je+1), T(isB:ie+1,jsB:je+1), and S(isB:ie+1,jsB:je+1).
- Parameters
[in] g: Ocean grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[in] h: Layer thickness [H ~> kg m-2][in] tv: Thermodynamic variables[out] pfu: Zonal acceleration [m s-2][out] pfv: Meridional acceleration [m s-2]cs: Finite volume PGF control structurep_atm: The pressure at the ice-ocean or atmosphere-ocean interface [Pa].[out] pbce: The baroclinic pressure anomaly in each layer due to eta anomalies [m2 s-2 H-1 ~> m s-2 or m4 s-2 kg-1].[out] eta: The bottom mass used to calculate PFu and PFv [H ~> m or kg m-2], with any tidal contributions or compressibility compensation.
-
subroutine, public mom_pressureforce_blk_afv::pressureforce_blk_afv_bouss(h h, tv tv, PFu PFu, PFv PFv, G G, GV GV, US US, CS CS, ALE_CSp ALE_CSp, p_atm p_atm, pbce pbce, eta eta) Boussinesq analytically-integrated finite volume form of pressure gradient.
Determines the acceleration due to hydrostatic pressure forces, using the finite volume form of the terms and analytic integrals in depth, making the Boussinesq approximation. This version uses code-blocking for threads.
To work, the following fields must be set outside of the usual (is:ie,js:je) range before this subroutine is called: h(isB:ie+1,jsB:je+1), T(isB:ie+1,jsB:je+1), and S(isB:ie+1,jsB:je+1).
- Parameters
[in] g: Ocean grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[in] h: Layer thickness [H ~> m or kg m-2][in] tv: Thermodynamic variables[out] pfu: Zonal acceleration [m s-2][out] pfv: Meridional acceleration [m s-2]cs: Finite volume PGF control structureale_csp: ALE control structurep_atm: The pressure at the ice-ocean or atmosphere-ocean interface [Pa].[out] pbce: The baroclinic pressure anomaly in each layer due to eta anomalies [m2 s-2 H-1 ~> m s-2 or m4 s-2 kg-1].[out] eta: The bottom mass used to calculate PFu and PFv [H ~> m or kg m-2], with any tidal contributions or compressibility compensation.
-
subroutine, public mom_pressureforce_blk_afv::pressureforce_blk_afv_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS, tides_CSp tides_CSp) Initializes the finite volume pressure gradient control structure.
- Parameters
[in] time: Current model time[in] g: Ocean grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: Parameter file handles[inout] diag: Diagnostics control structurecs: Finite volume PGF control structuretides_csp: Tides control structure
-
subroutine, public mom_pressureforce_blk_afv::pressureforce_blk_afv_end(CS CS) Deallocates the finite volume pressure gradient control structure.
- Parameters
cs: Blocked AFV pressure control structure that will be deallocated in this subroutine.
-
-
namespace
mom_pressureforce_mont¶ Provides the Montgomery potential form of pressure gradient.
Provides the Boussunesq and non-Boussinesq forms of the horizontal accelerations due to pressure gradients using the Montgomery potential. A second-order accurate, centered scheme is used. If a split time stepping scheme is used, the vertical decomposition into barotropic and baroclinic contributions described by Hallberg (J Comp Phys 1997) is used. With a nonlinear equation of state, compressibility is added along the lines proposed by Sun et al. (JPO 1999), but with compressibility coefficients based on a fit to a user-provided reference profile.
Unnamed Group
-
subroutine, public mom_pressureforce_mont::pressureforce_mont_nonbouss(h h, tv tv, PFu PFu, PFv PFv, G G, GV GV, US US, CS CS, p_atm p_atm, pbce pbce, eta eta) Non-Boussinesq Montgomery-potential form of pressure gradient.
Determines the acceleration due to pressure forces in a non-Boussinesq fluid using the compressibility compensated (if appropriate) Montgomery-potential form described in Hallberg (Ocean Mod., 2005).
To work, the following fields must be set outside of the usual (is:ie,js:je) range before this subroutine is called: h(isB:ie+1,jsB:je+1), T(isB:ie+1,jsB:je+1), and S(isB:ie+1,jsB:je+1).
- Parameters
[in] g: Ocean grid structure.[in] gv: Vertical grid structure.[in] us: A dimensional unit scaling type[in] h: Layer thickness, [H ~> kg m-2].[in] tv: Thermodynamic variables.[out] pfu: Zonal acceleration due to pressure gradients (equal to -dM/dx) [m s-2].[out] pfv: Meridional acceleration due to pressure gradients (equal to -dM/dy) [m s-2].cs: Control structure for Montgomery potential PGFp_atm: The pressure at the ice-ocean or atmosphere-ocean [Pa].[out] pbce: The baroclinic pressure anomaly in[out] eta: Free surface height [H ~> kg m-1].
-
subroutine, public mom_pressureforce_mont::pressureforce_mont_bouss(h h, tv tv, PFu PFu, PFv PFv, G G, GV GV, US US, CS CS, p_atm p_atm, pbce pbce, eta eta) Boussinesq Montgomery-potential form of pressure gradient.
Determines the acceleration due to pressure forces.
To work, the following fields must be set outside of the usual (is:ie,js:je) range before this subroutine is called: h(isB:ie+1,jsB:je+1), T(isB:ie+1,jsB:je+1), and S(isB:ie+1,jsB:je+1).
- Parameters
[in] g: Ocean grid structure.[in] gv: Vertical grid structure.[in] us: A dimensional unit scaling type[in] h: Layer thickness [H ~> m].[in] tv: Thermodynamic variables.[out] pfu: Zonal acceleration due to pressure gradients (equal to -dM/dx) [m s-2].[out] pfv: Meridional acceleration due to pressure gradients (equal to -dM/dy) [m s2].cs: Control structure for Montgomery potential PGFp_atm: The pressure at the ice-ocean or atmosphere-ocean [Pa].[out] pbce: The baroclinic pressure anomaly in each layer due to free surface height anomalies [m2 s-2 H-1 ~> m s-2].[out] eta: Free surface height [H ~> m].
-
subroutine, public mom_pressureforce_mont::set_pbce_bouss(e e, tv tv, G G, GV GV, US US, Rho0 Rho0, GFS_scale GFS_scale, pbce pbce, rho_star rho_star) Determines the partial derivative of the acceleration due to pressure forces with the free surface height.
- Parameters
[in] g: Ocean grid structure[in] gv: Vertical grid structure[in] e: Interface height [Z ~> m].[in] tv: Thermodynamic variables[in] us: A dimensional unit scaling type[in] rho0: The “Boussinesq” ocean density [kg m-3].[in] gfs_scale: Ratio between gravity applied to top interface and the gravitational acceleration of the planet [nondim]. Usually this ratio is 1.[out] pbce: The baroclinic pressure anomaly in each layer due[in] rho_star: The layer densities (maybe compressibility
-
subroutine, public mom_pressureforce_mont::set_pbce_nonbouss(p p, tv tv, G G, GV GV, US US, GFS_scale GFS_scale, pbce pbce, alpha_star alpha_star) Determines the partial derivative of the acceleration due to pressure forces with the column mass.
- Parameters
[in] g: Ocean grid structure[in] gv: Vertical grid structure[in] p: Interface pressures [Pa].[in] tv: Thermodynamic variables[in] us: A dimensional unit scaling type[in] gfs_scale: Ratio between gravity applied to top interface and the gravitational acceleration of the planet [nondim]. Usually this ratio is 1.[out] pbce: The baroclinic pressure anomaly in each layer due to free surface height anomalies [L2 H-1 T-2 ~> m4 kg-1 s-2].[in] alpha_star: The layer specific volumes (maybe compressibility compensated) [m3 kg-1].
-
subroutine, public mom_pressureforce_mont::pressureforce_mont_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS, tides_CSp tides_CSp) Initialize the Montgomery-potential form of PGF control structure.
- Parameters
[in] time: Current model time[in] g: ocean grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: Parameter file handles[inout] diag: Diagnostics control structurecs: Montgomery PGF control structuretides_csp: Tides control structure
-
subroutine, public mom_pressureforce_mont::pressureforce_mont_end(CS CS) Deallocates the Montgomery-potential form of PGF control structure.
- Parameters
cs: Control structure for Montgomery potential PGF
-
-
namespace
mom_regridding¶ Generates vertical grids as part of the ALE algorithm.
A vertical grid is defined solely by the cell thicknesses, \(h\). Most calculations in this module start with the coordinate at the bottom of the column set to -depth, and use a increasing value of coordinate with decreasing k. This is consistent with the rest of MOM6 that uses position, \(z\) which is a negative quantity for most of the ocean.
A change in grid is define through a change in position of the interfaces:
\[ z^n_{k+1/2} = z^{n-1}_{k+1/2} + \Delta z_{k+1/2} \]with the positive upward coordinate convention\[ z_{k-1/2} = z_{k+1/2} + h_k \]so that\[ h^n_k = h^{n-1}_k + ( \Delta z_{k-1/2} - \Delta z_{k+1/2} ) \]Original date of creation: 2008.06.09 by L. White
Functions
-
subroutine, public mom_regridding::initialize_regridding(CS CS, GV GV, US US, max_depth max_depth, param_file param_file, mdl mdl, coord_mode coord_mode, param_prefix param_prefix, param_suffix param_suffix) Initialization and configures a regridding control structure based on customizable run-time parameters.
- Parameters
[inout] cs: Regridding control structure[in] gv: Ocean vertical grid structure[in] us: A dimensional unit scaling type[in] max_depth: The maximum depth of the ocean [Z ~> m].[in] param_file: Parameter file[in] mdl: Name of calling module.[in] coord_mode: Coordinate mode[in] param_prefix: String to prefix to parameter names. If empty, causes main model parameters to be used.[in] param_suffix: String to append to parameter names.
-
subroutine
check_grid_def(filename filename, varname varname, expected_units expected_units, msg msg, ierr ierr)¶ Do some basic checks on the vertical grid definition file, variable.
- Parameters
[in] filename: File name[in] varname: Variable name[in] expected_units: Expected units of variable[inout] msg: Message to use for errors[out] ierr: True if an error occurs
-
subroutine, public mom_regridding::end_regridding(CS CS) Deallocation of regridding memory.
- Parameters
[inout] cs: Regridding control structure
-
subroutine, public mom_regridding::regridding_main(remapCS remapCS, CS CS, G G, GV GV, h h, tv tv, h_new h_new, dzInterface dzInterface, frac_shelf_h frac_shelf_h, conv_adjust conv_adjust) Dispatching regridding routine for orchestrating regridding & remapping.
- Parameters
[in] remapcs: Remapping parameters and options[in] cs: Regridding control structure[in] g: Ocean grid structure[in] gv: Ocean vertical grid structure[inout] h: Current 3D grid obtained after the last time step[inout] tv: Thermodynamical variables (T, S, …)[inout] h_new: New 3D grid consistent with target coordinate[inout] dzinterface: The change in position of each interfacefrac_shelf_h: Fractional ice shelf coverage[in] conv_adjust: If true, do convective adjustment
-
subroutine
calc_h_new_by_dz(CS CS, G G, GV GV, h h, dzInterface dzInterface, h_new h_new)¶ Calculates h_new from h + delta_k dzInterface.
- Parameters
[in] cs: Regridding control structure[in] g: Grid structure[in] gv: Ocean vertical grid structure[in] h: Old layer thicknesses (arbitrary units)[in] dzinterface: Change in interface positions (same as h)[inout] h_new: New layer thicknesses (same as h)
-
subroutine, public mom_regridding::check_remapping_grid(G G, GV GV, h h, dzInterface dzInterface, msg msg) Check that the total thickness of two grids match.
- Parameters
[in] g: Grid structure[in] gv: Ocean vertical grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][in] dzinterface: Change in interface positions [H ~> m or kg m-2][in] msg: Message to append to errors
-
subroutine, public mom_regridding::check_grid_column(nk nk, depth depth, h h, dzInterface dzInterface, msg msg) Check that the total thickness of new and old grids are consistent.
- Parameters
[in] nk: Number of cells[in] depth: Depth of bottom [Z ~> m] or arbitrary units[in] h: Cell thicknesses [Z ~> m] or arbitrary units[in] dzinterface: Change in interface positions (same units as h)[in] msg: Message to append to errors
-
subroutine
filtered_grid_motion(CS CS, nk nk, z_old z_old, z_new z_new, dz_g dz_g)¶ Returns the change in interface position motion after filtering and assuming the top and bottom interfaces do not move. The filtering is a function of depth, and is applied as the integrated average filtering over the trajectory of the interface. By design, this code can not give tangled interfaces provided that z_old and z_new are not already tangled.
- Parameters
[in] cs: Regridding control structure[in] nk: Number of cells in source grid[in] z_old: Old grid position [m][in] z_new: New grid position [m][inout] dz_g: Change in interface positions [m]
-
subroutine
build_zstar_grid(CS CS, G G, GV GV, h h, dzInterface dzInterface, frac_shelf_h frac_shelf_h)¶ Builds a z*-ccordinate grid with partial steps (Adcroft and Campin, 2004). z* is defined as z* = (z-eta)/(H+eta)*H s.t. z*=0 when z=eta and z*=-H when z=-H .
- Parameters
[in] cs: Regridding control structure[in] g: Ocean grid structure[in] gv: ocean vertical grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][inout] dzinterface: The change in interface depth [H ~> m or kg m-2].frac_shelf_h: Fractional ice shelf coverage.
-
subroutine
build_sigma_grid(CS CS, G G, GV GV, h h, dzInterface dzInterface)¶ This routine builds a grid based on terrain-following coordinates.
- Parameters
[in] cs: Regridding control structure[in] g: Ocean grid structure[in] gv: ocean vertical grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][inout] dzinterface: The change in interface depth [H ~> m or kg m-2]
-
subroutine
build_rho_grid(G G, GV GV, h h, tv tv, dzInterface dzInterface, remapCS remapCS, CS CS)¶ This routine builds a new grid based on a given set of target interface densities.
- Parameters
[in] g: Ocean grid structure[in] gv: Ocean vertical grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][in] tv: Thermodynamics structure[inout] dzinterface: The change in interface depth [H ~> m or kg m-2][in] remapcs: The remapping control structure[in] cs: Regridding control structure
-
subroutine
build_grid_hycom1(G G, GV GV, h h, tv tv, h_new h_new, dzInterface dzInterface, CS CS)¶ Builds a simple HyCOM-like grid with the deepest location of potential density interpolated from the column profile and a clipping of depth for each interface to a fixed z* or p* grid. This should probably be (optionally?) changed to find the nearest location of the target density.
- Remark
{ Based on Bleck, 2002: An oceanice general circulation model framed in hybrid isopycnic-Cartesian coordinates, Ocean Modelling 37, 55-88. http://dx.doi.org/10.1016/S1463-5003(01)00012-9 }
- Parameters
[in] g: Grid structure[in] gv: Ocean vertical grid structure[in] h: Existing model thickness [H ~> m or kg m-2][in] tv: Thermodynamics structure[in] cs: Regridding control structure[inout] h_new: New layer thicknesses [H ~> m or kg m-2][inout] dzinterface: Changes in interface position
-
subroutine
build_grid_adaptive(G G, GV GV, h h, tv tv, dzInterface dzInterface, remapCS remapCS, CS CS)¶ This subroutine builds an adaptive grid that follows density surfaces where possible, subject to constraints on the smoothness of interface heights.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][in] tv: A structure pointing to various thermodynamic variables[inout] dzinterface: The change in interface depth [H ~> m or kg m-2][in] remapcs: The remapping control structure[in] cs: Regridding control structure
-
subroutine
build_grid_slight(G G, GV GV, h h, tv tv, dzInterface dzInterface, CS CS)¶ Builds a grid that tracks density interfaces for water that is denser than the surface density plus an increment of some number of layers, and uses all lighter layers uniformly above this location. Note that this amounts to interpolating to find the depth of an arbitrary (non-integer) interface index which should make the results vary smoothly in space to the extent that the surface density and interior stratification vary smoothly in space. Over shallow topography, this will tend to give a uniform sigma-like coordinate. For sufficiently shallow water, a minimum grid spacing is used to avoid certain instabilities.
- Parameters
[in] g: Grid structure[in] gv: Ocean vertical grid structure[in] h: Existing model thickness [H ~> m or kg m-2][in] tv: Thermodynamics structure[inout] dzinterface: Changes in interface position[in] cs: Regridding control structure
-
subroutine
adjust_interface_motion(CS CS, nk nk, h_old h_old, dz_int dz_int)¶ Adjust dz_Interface to ensure non-negative future thicknesses.
- Parameters
[in] cs: Regridding control structure[in] nk: Number of layers in h_old[in] h_old: Minium allowed thickness of h [H ~> m or kg m-2][inout] dz_int: Minium allowed thickness of h [H ~> m or kg m-2]
-
subroutine
build_grid_arbitrary(G G, GV GV, h h, dzInterface dzInterface, h_new h_new, CS CS)¶ - Parameters
[in] g: Ocean grid structure[in] gv: Ocean vertical grid structure[in] h: Original layer thicknesses [H ~> m or kg m-2][inout] dzinterface: The change in interface depth [H ~> m or kg m-2][inout] h_new: New layer thicknesses [H ~> m or kg m-2][in] cs: Regridding control structure
-
subroutine, public mom_regridding::inflate_vanished_layers_old(CS CS, G G, GV GV, h h) - Parameters
[in] cs: Regridding control structure[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[inout] h: Layer thicknesses [H ~> m or kg m-2]
-
subroutine
convective_adjustment(G G, GV GV, h h, tv tv)¶ Achieve convective adjustment by swapping layers.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[inout] h: Layer thicknesses [H ~> m or kg m-2][inout] tv: A structure pointing to various thermodynamic variables
-
real function, dimension(nk), public mom_regridding::uniformresolution(nk nk, coordMode coordMode, maxDepth maxDepth, rhoLight rhoLight, rhoHeavy rhoHeavy) Return a uniform resolution vector in the units of the coordinate.
- Return
The returned uniform resolution grid.
- Parameters
[in] nk: Number of cells in source grid[in] coordmode: A string indicating the coordinate mode. See the documenttion for regrid_consts for the recognized values.[in] maxdepth: The range of the grid values in some modes[in] rholight: The minimum value of the grid in RHO mode[in] rhoheavy: The maximum value of the grid in RHO mode
-
subroutine
initcoord(CS CS, GV GV, coord_mode coord_mode)¶ Initialize the coordinate resolutions by calling the appropriate initialization routine for the specified coordinate mode.
- Parameters
[inout] cs: Regridding control structure[in] coord_mode: A string indicating the coordinate mode. See the documenttion for regrid_consts for the recognized values.[in] gv: Ocean vertical grid structure
-
subroutine, public mom_regridding::setcoordinateresolution(dz dz, CS CS, scale scale) Set the fixed resolution data.
- Parameters
[in] dz: A vector of vertical grid spacings[inout] cs: Regridding control structure[in] scale: A scaling factor converting dz to coordRes
-
subroutine, public mom_regridding::set_target_densities_from_gv(GV GV, CS CS) Set target densities based on the old Rlay variable.
- Parameters
[in] gv: Ocean vertical grid structure[inout] cs: Regridding control structure
-
subroutine, public mom_regridding::set_target_densities(CS CS, rho_int rho_int) Set target densities based on vector of interface values.
- Parameters
[inout] cs: Regridding control structure[in] rho_int: Interface densities
-
subroutine, public mom_regridding::set_regrid_max_depths(CS CS, max_depths max_depths, units_to_H units_to_H) Set maximum interface depths based on a vector of input values.
- Parameters
[inout] cs: Regridding control structure[in] max_depths: Maximum interface depths, in arbitrary units[in] units_to_h: A conversion factor for max_depths into H units
-
subroutine, public mom_regridding::set_regrid_max_thickness(CS CS, max_h max_h, units_to_H units_to_H) Set maximum layer thicknesses based on a vector of input values.
- Parameters
[inout] cs: Regridding control structure[in] max_h: Maximum interface depths, in arbitrary units[in] units_to_h: A conversion factor for max_h into H units
-
real function, dimension(cs%nk), public mom_regridding::getcoordinateresolution(CS CS, undo_scaling undo_scaling) Query the fixed resolution data.
- Parameters
[in] cs: Regridding control structure[in] undo_scaling: If present and true, undo any internal rescaling of the resolution data.
-
real function, dimension(cs%nk+1), public mom_regridding::getcoordinateinterfaces(CS CS, undo_scaling undo_scaling) Query the target coordinate interface positions.
- Return
Interface positions in target coordinate
- Parameters
[in] cs: Regridding control structure[in] undo_scaling: If present and true, undo any internal rescaling of the resolution data.
-
character(len=20) function, public mom_regridding::getcoordinateunits(CS CS) Query the target coordinate units.
- Parameters
[in] cs: Regridding control structure
-
character(len=20) function, public mom_regridding::getcoordinateshortname(CS CS) Query the short name of the coordinate.
- Parameters
[in] cs: Regridding control structure
-
subroutine, public mom_regridding::set_regrid_params(CS CS, boundary_extrapolation boundary_extrapolation, min_thickness min_thickness, old_grid_weight old_grid_weight, interp_scheme interp_scheme, depth_of_time_filter_shallow depth_of_time_filter_shallow, depth_of_time_filter_deep depth_of_time_filter_deep, compress_fraction compress_fraction, dz_min_surface dz_min_surface, nz_fixed_surface nz_fixed_surface, Rho_ML_avg_depth Rho_ML_avg_depth, nlay_ML_to_interior nlay_ML_to_interior, fix_haloclines fix_haloclines, halocline_filt_len halocline_filt_len, halocline_strat_tol halocline_strat_tol, integrate_downward_for_e integrate_downward_for_e, adaptTimeRatio adaptTimeRatio, adaptZoom adaptZoom, adaptZoomCoeff adaptZoomCoeff, adaptBuoyCoeff adaptBuoyCoeff, adaptAlpha adaptAlpha, adaptDoMin adaptDoMin) Can be used to set any of the parameters for MOM_regridding.
- Parameters
[inout] cs: Regridding control structure[in] boundary_extrapolation: Extrapolate in boundary cells[in] min_thickness: Minimum thickness allowed when building the new grid [H ~> m or kg m-2][in] old_grid_weight: Weight given to old coordinate when time-filtering grid[in] interp_scheme: Interpolation method for state-dependent coordinates[in] depth_of_time_filter_shallow: Depth to start cubic [H ~> m or kg m-2][in] depth_of_time_filter_deep: Depth to end cubic [H ~> m or kg m-2][in] compress_fraction: Fraction of compressibility to add to potential density[in] dz_min_surface: The fixed resolution in the topmost SLight_nkml_min layers [H ~> m or kg m-2][in] nz_fixed_surface: The number of fixed-thickness layers at the top of the model[in] rho_ml_avg_depth: Averaging depth over which to determine mixed layer potential density [H ~> m or kg m-2][in] nlay_ml_to_interior: Number of layers to offset the mixed layer density to find resolved stratification [nondim][in] fix_haloclines: Detect regions with much weaker stratification in the coordinate[in] halocline_filt_len: Length scale over which to filter T & S when looking for spuriously unstable water mass profiles [m][in] halocline_strat_tol: Value of the stratification ratio that defines a problematic halocline region.[in] integrate_downward_for_e: If true, integrate for interface positions downward from the top.[in] adapttimeratio: Ratio of the ALE timestep to the grid timescale [nondim].[in] adaptzoom: Depth of near-surface zooming region [H ~> m or kg m-2].[in] adaptzoomcoeff: Coefficient of near-surface zooming diffusivity [nondim].[in] adaptbuoycoeff: Coefficient of buoyancy diffusivity [nondim].[in] adaptalpha: Scaling factor on optimization tendency [nondim].[in] adaptdomin: If true, make a HyCOM-like mixed layer by preventing interfaces from being shallower than the depths specified by the regridding coordinate.
-
integer function, public mom_regridding::get_regrid_size(CS CS) Returns the number of levels/layers in the regridding control structure.
- Parameters
[inout] cs: Regridding control structure
-
type(zlike_cs) function, public mom_regridding::get_zlike_cs(CS CS) This returns a copy of the zlike_CS stored in the regridding control structure.
- Parameters
[in] cs: Regridding control structure
-
type(sigma_cs) function, public mom_regridding::get_sigma_cs(CS CS) This returns a copy of the sigma_CS stored in the regridding control structure.
- Parameters
[in] cs: Regridding control structure
-
type(rho_cs) function, public mom_regridding::get_rho_cs(CS CS) This returns a copy of the rho_CS stored in the regridding control structure.
- Parameters
[in] cs: Regridding control structure
-
real function, dimension(cs%nk), public mom_regridding::getstaticthickness(CS CS, SSH SSH, depth depth) Return coordinate-derived thicknesses for fixed coordinate systems.
- Return
The returned thicknesses in the units of depth
- Parameters
[in] cs: Regridding control structure[in] ssh: The sea surface height, in the same units as depth[in] depth: The maximum depth of the grid, often [Z ~> m]
-
subroutine
dz_function1(string string, dz dz)¶ Parses a string and generates a dz(:) profile that goes like k**power.
- Parameters
[in] string: String with list of parameters in form dz_min, H_total, power, precision[inout] dz: Profile of nominal thicknesses
-
integer function mom_regridding::rho_function1(string string, rho_target rho_target) Parses a string and generates a rho_target(:) profile with refined resolution downward and returns the number of levels.
- Parameters
[in] string: String with list of parameters in form dz_min, H_total, power, precision[inout] rho_target: Profile of interface densities
Variables
-
character(len=*), parameter, public mom_regridding::regriddingcoordinatemodedoc = " LAYER - Isopycnal or stacked shallow water layersn"// " ZSTAR, Z* - stretched geopotential z*n"// " SIGMA_SHELF_ZSTAR - stretched geopotential z* ignoring shelfn"// " SIGMA - terrain following coordinatesn"// " RHO - continuous isopycnaln"// " HYCOM1 - HyCOM-like hybrid coordinaten"// " SLIGHT - stretched coordinates above continuous isopycnaln"// " ADAPTIVE - optimize for smooth neutral density surfaces" Documentation for coordinate options.
-
character(len=*), parameter, public mom_regridding::regriddinginterpschemedoc = " P1M_H2 (2nd-order accurate)n"// " P1M_H4 (2nd-order accurate)n"// " P1M_IH4 (2nd-order accurate)n"// " PLM (2nd-order accurate)n"// " PPM_H4 (3rd-order accurate)n"// " PPM_IH4 (3rd-order accurate)n"// " P3M_IH4IH3 (4th-order accurate)n"// " P3M_IH6IH5 (4th-order accurate)n"// " PQM_IH4IH3 (4th-order accurate)n"// " PQM_IH6IH5 (5th-order accurate)" Documentation for regridding interpolation schemes.
-
character(len=*), parameter, public mom_regridding::regriddingdefaultinterpscheme = "P1M_H2" Default interpolation scheme.
-
logical, parameter, public mom_regridding::regriddingdefaultboundaryextrapolation = .false. Default mode for boundary extrapolation.
-
real, parameter, public mom_regridding::regriddingdefaultminthickness = 1.e-3 Default minimum thickness for some coordinate generation modes.
-
-
namespace
mom_regularize_layers¶ Provides regularization of layers in isopycnal mode.
Unnamed Group
-
integer
id_clock_pass¶ Clock IDs.
-
integer
id_clock_eos¶ Clock IDs.
-
subroutine, public mom_regularize_layers::regularize_layers(h h, tv tv, dt dt, ea ea, eb eb, G G, GV GV, CS CS) This subroutine partially steps the bulk mixed layer model. The following processes are executed, in the order listed.
- Parameters
[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[inout] h: Layer thicknesses [H ~> m or kg m-2].[inout] tv: A structure containing pointers to any available thermodynamic fields. Absent fields have NULL ptrs.[in] dt: Time increment [s].[inout] ea: The amount of fluid moved downward into a[inout] eb: The amount of fluid moved upward into a layercs: The control structure returned by a previous call to regularize_layers_init.
-
subroutine
regularize_surface(h h, tv tv, dt dt, ea ea, eb eb, G G, GV GV, CS CS)¶ This subroutine ensures that there is a degree of horizontal smoothness in the depths of the near-surface interfaces.
- Parameters
[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[inout] h: Layer thicknesses [H ~> m or kg m-2].[inout] tv: A structure containing pointers to any available thermodynamic fields. Absent fields have NULL ptrs.[in] dt: Time increment [s].[inout] ea: The amount of fluid moved downward into a[inout] eb: The amount of fluid moved upward into a layercs: The control structure returned by a previous call to regularize_layers_init.
-
subroutine
find_deficit_ratios(e e, def_rat_u def_rat_u, def_rat_v def_rat_v, G G, GV GV, CS CS, def_rat_u_2lay def_rat_u_2lay, def_rat_v_2lay def_rat_v_2lay, halo halo, h h)¶ This subroutine determines the amount by which the harmonic mean thickness at velocity points differ from the arithmetic means, relative to the the arithmetic means, after eliminating thickness variations that are solely due to topography and aggregating all interior layers into one.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] e: Interface depths [H ~> m or kg m-2][out] def_rat_u: The thickness deficit ratio at u points,[out] def_rat_v: The thickness deficit ratio at v points,cs: The control structure returned by a previous call to regularize_layers_init.[out] def_rat_u_2lay: The thickness deficit ratio at u[out] def_rat_v_2lay: The thickness deficit ratio at v[in] halo: An extra-wide halo size, 0 by default.[in] h: Layer thicknesses [H ~> m or kg m-2].
-
subroutine, public mom_regularize_layers::regularize_layers_init(Time Time, G G, GV GV, param_file param_file, diag diag, CS CS) Initializes the regularize_layers control structure.
- Parameters
[in] time: The current model time.[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] param_file: A structure to parse for run-time parameters.[inout] diag: A structure that is used to regulate diagnostic output.cs: A pointer that is set to point to the control structure for this module.
-
integer
-
namespace
mom_remapping¶ Provides column-wise vertical remapping functions.
Functions
-
subroutine, public mom_remapping::remapping_set_param(CS CS, remapping_scheme remapping_scheme, boundary_extrapolation boundary_extrapolation, check_reconstruction check_reconstruction, check_remapping check_remapping, force_bounds_in_subcell force_bounds_in_subcell) Set parameters within remapping object.
- Parameters
[inout] cs: Remapping control structure[in] remapping_scheme: Remapping scheme to use[in] boundary_extrapolation: Indicate to extrapolate in boundary cells[in] check_reconstruction: Indicate to check reconstructions[in] check_remapping: Indicate to check results of remapping[in] force_bounds_in_subcell: Force subcells values to be bounded
-
subroutine, public mom_remapping::extract_member_remapping_cs(CS CS, remapping_scheme remapping_scheme, degree degree, boundary_extrapolation boundary_extrapolation, check_reconstruction check_reconstruction, check_remapping check_remapping, force_bounds_in_subcell force_bounds_in_subcell) - Parameters
[in] cs: Control structure for remapping module[out] remapping_scheme: Determines which reconstruction scheme to use[out] degree: Degree of polynomial reconstruction[out] boundary_extrapolation: If true, extrapolate boundaries[out] check_reconstruction: If true, reconstructions are checked for consistency.[out] check_remapping: If true, the result of remapping are checked for conservation and bounds.[out] force_bounds_in_subcell: If true, the intermediate values used in remapping are forced to be bounded.
-
subroutine
buildgridfromh(nz nz, h h, x x)¶ Calculate edge coordinate x from cell width h.
- Parameters
[in] nz: Number of cells[in] h: Cell widths[inout] x: Edge coordiantes starting at x(1)=0
-
logical function mom_remapping::ispossumerrsignificant(n1 n1, sum1 sum1, n2 n2, sum2 sum2) Compare two summation estimates of positive data and judge if due to more than round-off. When two sums are calculated from different vectors that should add up to the same value, the results can differ by round off. The round off error can be bounded to be proportional to the number of operations. This function returns true if the difference between sum1 and sum2 is larger than than the estimated round off bound.
- Note
This estimate/function is only valid for summation of positive data.
- Return
True if difference in sums is large
- Parameters
[in] n1: Number of values in sum1[in] n2: Number of values in sum2[in] sum1: Sum of n1 values[in] sum2: Sum of n2 values
-
subroutine, public mom_remapping::remapping_core_h(CS CS, n0 n0, h0 h0, u0 u0, n1 n1, h1 h1, u1 u1, h_neglect h_neglect, h_neglect_edge h_neglect_edge) Remaps column of values u0 on grid h0 to grid h1 assuming the top edge is aligned.
- Parameters
[in] cs: Remapping control structure[in] n0: Number of cells on source grid[in] h0: Cell widths on source grid[in] u0: Cell averages on source grid[in] n1: Number of cells on target grid[in] h1: Cell widths on target grid[out] u1: Cell averages on target grid[in] h_neglect: A negligibly small width for the purpose of cell reconstructions in the same units as h0.[in] h_neglect_edge: A negligibly small width for the purpose of edge value calculations in the same units as h0.
-
subroutine, public mom_remapping::remapping_core_w(CS CS, n0 n0, h0 h0, u0 u0, n1 n1, dx dx, u1 u1, h_neglect h_neglect, h_neglect_edge h_neglect_edge) Remaps column of values u0 on grid h0 to implied grid h1 where the interfaces of h1 differ from those of h0 by dx.
- Parameters
[in] cs: Remapping control structure[in] n0: Number of cells on source grid[in] h0: Cell widths on source grid[in] u0: Cell averages on source grid[in] n1: Number of cells on target grid[in] dx: Cell widths on target grid[out] u1: Cell averages on target grid[in] h_neglect: A negligibly small width for the purpose of cell reconstructions in the same units as h0.[in] h_neglect_edge: A negligibly small width for the purpose of edge value calculations in the same units as h0.
-
subroutine, public mom_remapping::build_reconstructions_1d(CS CS, n0 n0, h0 h0, u0 u0, ppoly_r_coefs ppoly_r_coefs, ppoly_r_E ppoly_r_E, ppoly_r_S ppoly_r_S, iMethod iMethod, h_neglect h_neglect, h_neglect_edge h_neglect_edge) Creates polynomial reconstructions of u0 on the source grid h0.
- Parameters
[in] cs: Remapping control structure[in] n0: Number of cells on source grid[in] h0: Cell widths on source grid[in] u0: Cell averages on source grid[out] ppoly_r_coefs: Coefficients of polynomial[out] ppoly_r_e: Edge value of polynomial[out] ppoly_r_s: Edge slope of polynomial[out] imethod: Integration method[in] h_neglect: A negligibly small width for the purpose of cell reconstructions in the same units as h0.[in] h_neglect_edge: A negligibly small width for the purpose of edge value calculations in the same units as h0.
-
subroutine
check_reconstructions_1d(n0 n0, h0 h0, u0 u0, deg deg, boundary_extrapolation boundary_extrapolation, ppoly_r_coefs ppoly_r_coefs, ppoly_r_E ppoly_r_E, ppoly_r_S ppoly_r_S)¶ Checks that edge values and reconstructions satisfy bounds.
- Parameters
[in] n0: Number of cells on source grid[in] h0: Cell widths on source grid[in] u0: Cell averages on source grid[in] deg: Degree of polynomial reconstruction[in] boundary_extrapolation: Extrapolate at boundaries if true[out] ppoly_r_coefs: Coefficients of polynomial[out] ppoly_r_e: Edge value of polynomial[out] ppoly_r_s: Edge slope of polynomial
-
subroutine
remap_via_sub_cells(n0 n0, h0 h0, u0 u0, ppoly0_E ppoly0_E, ppoly0_coefs ppoly0_coefs, n1 n1, h1 h1, method method, force_bounds_in_subcell force_bounds_in_subcell, u1 u1, uh_err uh_err, ah_sub ah_sub, aisub_src aisub_src, aiss aiss, aise aise)¶ Remaps column of n0 values u0 on grid h0 to grid h1 with n1 cells by calculating the n0+n1+1 sub-integrals of the intersection of h0 and h1, and the summing the appropriate integrals into the h1*u1 values. h0 and h1 must have the same units.
- Parameters
[in] n0: Number of cells in source grid[in] h0: Source grid widths (size n0)[in] u0: Source cell averages (size n0)[in] ppoly0_e: Edge value of polynomial[in] ppoly0_coefs: Coefficients of polynomial[in] n1: Number of cells in target grid[in] h1: Target grid widths (size n1)[in] method: Remapping scheme to use[in] force_bounds_in_subcell: Force sub-cell values to be bounded[out] u1: Target cell averages (size n1)[out] uh_err: Estimate of bound on error in sum of u*h[out] ah_sub: h_sub[out] aisub_src: i_sub_src[out] aiss: isrc_start[out] aise: isrc_ens
-
real function, public mom_remapping::average_value_ppoly(n0 n0, u0 u0, ppoly0_E ppoly0_E, ppoly0_coefs ppoly0_coefs, method method, i0 i0, xa xa, xb xb) Returns the average value of a reconstruction within a single source cell, i0, between the non-dimensional positions xa and xb (xa<=xb) with dimensional separation dh.
- Parameters
[in] n0: Number of cells in source grid[in] u0: Cell means[in] ppoly0_e: Edge value of polynomial[in] ppoly0_coefs: Coefficients of polynomial[in] method: Remapping scheme to use[in] i0: Source cell index[in] xa: Non-dimensional start position within source cell[in] xb: Non-dimensional end position within source cell
-
subroutine
measure_input_bounds(n0 n0, h0 h0, u0 u0, ppoly_E ppoly_E, h0tot h0tot, h0err h0err, u0tot u0tot, u0err u0err, u0min u0min, u0max u0max)¶ Measure totals and bounds on source grid.
- Parameters
[in] n0: Number of cells on source grid[in] h0: Cell widths on source grid[in] u0: Cell averages on source grid[in] ppoly_e: Cell edge values on source grid[out] h0tot: Sum of cell widths[out] h0err: Magnitude of round-off error in h0tot[out] u0tot: Sum of cell widths times values[out] u0err: Magnitude of round-off error in u0tot[out] u0min: Minimum value in reconstructions of u0[out] u0max: Maximum value in reconstructions of u0
-
subroutine
measure_output_bounds(n1 n1, h1 h1, u1 u1, h1tot h1tot, h1err h1err, u1tot u1tot, u1err u1err, u1min u1min, u1max u1max)¶ Measure totals and bounds on destination grid.
- Parameters
[in] n1: Number of cells on destination grid[in] h1: Cell widths on destination grid[in] u1: Cell averages on destination grid[out] h1tot: Sum of cell widths[out] h1err: Magnitude of round-off error in h1tot[out] u1tot: Sum of cell widths times values[out] u1err: Magnitude of round-off error in u1tot[out] u1min: Minimum value in reconstructions of u1[out] u1max: Maximum value in reconstructions of u1
-
subroutine
remapbyprojection(n0 n0, h0 h0, u0 u0, ppoly0_E ppoly0_E, ppoly0_coefs ppoly0_coefs, n1 n1, h1 h1, method method, u1 u1, h_neglect h_neglect)¶ Remaps column of values u0 on grid h0 to grid h1 by integrating over the projection of each h1 cell onto the h0 grid.
- Parameters
[in] n0: Number of cells in source grid[in] h0: Source grid widths (size n0)[in] u0: Source cell averages (size n0)[in] ppoly0_e: Edge value of polynomial[in] ppoly0_coefs: Coefficients of polynomial[in] n1: Number of cells in target grid[in] h1: Target grid widths (size n1)[in] method: Remapping scheme to use[out] u1: Target cell averages (size n1)[in] h_neglect: A negligibly small width for the purpose of cell reconstructions in the same units as h.
-
subroutine
remapbydeltaz(n0 n0, h0 h0, u0 u0, ppoly0_E ppoly0_E, ppoly0_coefs ppoly0_coefs, n1 n1, dx1 dx1, method method, u1 u1, h1 h1, h_neglect h_neglect)¶ Remaps column of values u0 on grid h0 to implied grid h1 where the interfaces of h1 differ from those of h0 by dx. The new grid is defined relative to the original grid by change dx1(:) = xNew(:) - xOld(:) and the remapping calculated so that hNew(k) qNew(k) = hOld(k) qOld(k) + F(k+1) - F(k) where F(k) = dx1(k) qAverage and where qAverage is the average qOld in the region zOld(k) to zNew(k).
- Parameters
[in] n0: Number of cells in source grid[in] h0: Source grid sizes (size n0)[in] u0: Source cell averages (size n0)[in] ppoly0_e: Edge value of polynomial[in] ppoly0_coefs: Coefficients of polynomial[in] n1: Number of cells in target grid[in] dx1: Target grid edge positions (size n1+1)[in] method: Remapping scheme to use[out] u1: Target cell averages (size n1)[out] h1: Target grid widths (size n1)[in] h_neglect: A negligibly small width for the purpose of cell reconstructions in the same units as h.
-
subroutine
integraterecononinterval(n0 n0, h0 h0, u0 u0, ppoly0_E ppoly0_E, ppoly0_coefs ppoly0_coefs, method method, xL xL, xR xR, hC hC, uAve uAve, jStart jStart, xStart xStart, h_neglect h_neglect)¶ Integrate the reconstructed column profile over a single cell.
- Parameters
[in] n0: Number of cells in source grid[in] h0: Source grid sizes (size n0)[in] u0: Source cell averages[in] ppoly0_e: Edge value of polynomial[in] ppoly0_coefs: Coefficients of polynomial[in] method: Remapping scheme to use[in] xl: Left edges of target cell[in] xr: Right edges of target cell[in] hc: Cell width hC = xR - xL[out] uave: Average value on target cell[inout] jstart: The index of the cell to start searching from On exit, contains index of last cell used[inout] xstart: The left edge position of cell jStart On first entry should be 0.[in] h_neglect: A negligibly small width for the purpose of cell reconstructions in the same units as h.
-
subroutine, public mom_remapping::dzfromh1h2(n1 n1, h1 h1, n2 n2, h2 h2, dx dx) Calculates the change in interface positions based on h1 and h2.
- Parameters
[in] n1: Number of cells on source grid[in] h1: Cell widths of source grid (size n1)[in] n2: Number of cells on target grid[in] h2: Cell widths of target grid (size n2)[out] dx: Change in interface position (size n2+1)
-
subroutine, public mom_remapping::initialize_remapping(CS CS, remapping_scheme remapping_scheme, boundary_extrapolation boundary_extrapolation, check_reconstruction check_reconstruction, check_remapping check_remapping, force_bounds_in_subcell force_bounds_in_subcell) Constructor for remapping control structure.
- Parameters
[inout] cs: Remapping control structure[in] remapping_scheme: Remapping scheme to use[in] boundary_extrapolation: Indicate to extrapolate in boundary cells[in] check_reconstruction: Indicate to check reconstructions[in] check_remapping: Indicate to check results of remapping[in] force_bounds_in_subcell: Force subcells values to be bounded
-
subroutine
setreconstructiontype(string string, CS CS)¶ Changes the method of reconstruction Use this routine to parse a string parameter specifying the reconstruction and re-allocates work arrays appropriately. It is called from initialize_remapping but can be called from an external module too.
- Parameters
[in] string: String to parse for method[inout] cs: Remapping control structure
-
subroutine, public mom_remapping::end_remapping(CS CS) Destrcutor for remapping control structure.
- Parameters
[inout] cs: Remapping control structure
-
logical function, public mom_remapping::remapping_unit_tests(verbose verbose) Runs unit tests on remapping functions. Should only be called from a single/root thread Returns True if a test fails, otherwise False.
- Parameters
[in] verbose: If true, write results to stdout
-
logical function mom_remapping::test_answer(verbose verbose, n n, u u, u_true u_true, label label) Returns true if any cell of u and u_true are not identical. Returns false otherwise.
- Parameters
[in] verbose: If true, write results to stdout[in] n: Number of cells in u[in] u: Values to test[in] u_true: Values to test against (correct answer)[in] label: Message
Variables
-
integer, parameter mom_remapping::remapping_pcm = 0 O(h^1) remapping scheme.
-
integer, parameter mom_remapping::remapping_plm = 1 O(h^2) remapping scheme.
-
integer, parameter mom_remapping::remapping_ppm_h4 = 2 O(h^3) remapping scheme.
-
integer, parameter mom_remapping::remapping_ppm_ih4 = 3 O(h^3) remapping scheme.
-
integer, parameter mom_remapping::remapping_pqm_ih4ih3 = 4 O(h^4) remapping scheme.
-
integer, parameter mom_remapping::remapping_pqm_ih6ih5 = 5 O(h^5) remapping scheme.
-
integer, parameter mom_remapping::integration_pcm = 0 Piecewise Constant Method.
-
integer, parameter mom_remapping::integration_plm = 1 Piecewise Linear Method.
-
integer, parameter mom_remapping::integration_ppm = 3 Piecewise Parabolic Method.
-
integer, parameter mom_remapping::integration_pqm = 5 Piecewise Quartic Method.
-
character(len=40) mom_remapping::mdl = "MOM_remapping" This module’s name.
-
character(len=256), public mom_remapping::remappingschemesdoc = "PCM (1st-order accurate)n"// "PLM (2nd-order accurate)n"// "PPM_H4 (3rd-order accurate)n"// "PPM_IH4 (3rd-order accurate)n"// "PQM_IH4IH3 (4th-order accurate)n"// "PQM_IH6IH5 (5th-order accurate)n" Documentation for external callers.
-
character(len=3), public mom_remapping::remappingdefaultscheme = "PLM" Default remapping method.
-
real, parameter mom_remapping::hneglect_dflt = 1.E-30 A thickness [H ~> m or kg m-2] that can be added to thicknesses in a denominator without changing the numerical result, except where a division by zero would otherwise occur.
-
logical, parameter mom_remapping::old_algorithm = .false. Use the old “broken” algorithm. This is a temporary measure to assist debugging until we delete the old algorithm.
-
-
namespace
mom_restart¶ The MOM6 facility for reading and writing restart files, and querying what has been read.
Unnamed Group
-
subroutine, public mom_restart::register_restart_field_as_obsolete(field_name field_name, replacement_name replacement_name, CS CS) - Parameters
[in] field_name: Name of restart field that is no longer in use[in] replacement_name: Name of replacement restart field, if applicablecs: A pointer to a MOM_restart_CS object (intent in/out)
-
subroutine
register_restart_field_ptr3d(f_ptr f_ptr, var_desc var_desc, mandatory mandatory, CS CS)¶ Register a 3-d field for restarts, providing the metadata in a structure.
- Parameters
[in] f_ptr: A pointer to the field to be read or written[in] var_desc: A structure with metadata about this variable[in] mandatory: If true, the run will abort if this field is not successfully read from the restart file.cs: A pointer to a MOM_restart_CS object (intent in/out)
-
subroutine
register_restart_field_ptr4d(f_ptr f_ptr, var_desc var_desc, mandatory mandatory, CS CS)¶ Register a 4-d field for restarts, providing the metadata in a structure.
- Parameters
[in] f_ptr: A pointer to the field to be read or written[in] var_desc: A structure with metadata about this variable[in] mandatory: If true, the run will abort if this field is not successfully read from the restart file.cs: A pointer to a MOM_restart_CS object (intent in/out)
-
subroutine
register_restart_field_ptr2d(f_ptr f_ptr, var_desc var_desc, mandatory mandatory, CS CS)¶ Register a 2-d field for restarts, providing the metadata in a structure.
- Parameters
[in] f_ptr: A pointer to the field to be read or written[in] var_desc: A structure with metadata about this variable[in] mandatory: If true, the run will abort if this field is not successfully read from the restart file.cs: A pointer to a MOM_restart_CS object (intent in/out)
-
subroutine
register_restart_field_ptr1d(f_ptr f_ptr, var_desc var_desc, mandatory mandatory, CS CS)¶ Register a 1-d field for restarts, providing the metadata in a structure.
- Parameters
[in] f_ptr: A pointer to the field to be read or written[in] var_desc: A structure with metadata about this variable[in] mandatory: If true, the run will abort if this field is not successfully read from the restart file.cs: A pointer to a MOM_restart_CS object (intent in/out)
-
subroutine
register_restart_field_ptr0d(f_ptr f_ptr, var_desc var_desc, mandatory mandatory, CS CS)¶ Register a 0-d field for restarts, providing the metadata in a structure.
- Parameters
[in] f_ptr: A pointer to the field to be read or written[in] var_desc: A structure with metadata about this variable[in] mandatory: If true, the run will abort if this field is not successfully read from the restart file.cs: A pointer to a MOM_restart_CS object (intent in/out)
-
subroutine
register_restart_field_4d(f_ptr f_ptr, name name, mandatory mandatory, CS CS, longname longname, units units, hor_grid hor_grid, z_grid z_grid, t_grid t_grid)¶ Register a 4-d field for restarts, providing the metadata as individual arguments.
- Parameters
[in] f_ptr: A pointer to the field to be read or written[in] name: variable name to be used in the restart file[in] mandatory: If true, the run will abort if this field is not successfully read from the restart file.cs: A pointer to a MOM_restart_CS object (intent in/out)[in] longname: variable long name[in] units: variable units[in] hor_grid: variable horizonal staggering, ‘h’ if absent[in] z_grid: variable vertical staggering, ‘L’ if absent[in] t_grid: time description: s, p, or 1, ‘s’ if absent
-
subroutine
register_restart_field_3d(f_ptr f_ptr, name name, mandatory mandatory, CS CS, longname longname, units units, hor_grid hor_grid, z_grid z_grid, t_grid t_grid)¶ Register a 3-d field for restarts, providing the metadata as individual arguments.
- Parameters
[in] f_ptr: A pointer to the field to be read or written[in] name: variable name to be used in the restart file[in] mandatory: If true, the run will abort if this field is not successfully read from the restart file.cs: A pointer to a MOM_restart_CS object (intent in/out)[in] longname: variable long name[in] units: variable units[in] hor_grid: variable horizonal staggering, ‘h’ if absent[in] z_grid: variable vertical staggering, ‘L’ if absent[in] t_grid: time description: s, p, or 1, ‘s’ if absent
-
subroutine
register_restart_field_2d(f_ptr f_ptr, name name, mandatory mandatory, CS CS, longname longname, units units, hor_grid hor_grid, z_grid z_grid, t_grid t_grid)¶ Register a 2-d field for restarts, providing the metadata as individual arguments.
- Parameters
[in] f_ptr: A pointer to the field to be read or written[in] name: variable name to be used in the restart file[in] mandatory: If true, the run will abort if this field is not successfully read from the restart file.cs: A pointer to a MOM_restart_CS object (intent in/out)[in] longname: variable long name[in] units: variable units[in] hor_grid: variable horizonal staggering, ‘h’ if absent[in] z_grid: variable vertical staggering, ‘1’ if absent[in] t_grid: time description: s, p, or 1, ‘s’ if absent
-
subroutine
register_restart_field_1d(f_ptr f_ptr, name name, mandatory mandatory, CS CS, longname longname, units units, hor_grid hor_grid, z_grid z_grid, t_grid t_grid)¶ Register a 1-d field for restarts, providing the metadata as individual arguments.
- Parameters
[in] f_ptr: A pointer to the field to be read or written[in] name: variable name to be used in the restart file[in] mandatory: If true, the run will abort if this field is not successfully read from the restart file.cs: A pointer to a MOM_restart_CS object (intent in/out)[in] longname: variable long name[in] units: variable units[in] hor_grid: variable horizonal staggering, ‘1’ if absent[in] z_grid: variable vertical staggering, ‘L’ if absent[in] t_grid: time description: s, p, or 1, ‘s’ if absent
-
subroutine
register_restart_field_0d(f_ptr f_ptr, name name, mandatory mandatory, CS CS, longname longname, units units, t_grid t_grid)¶ Register a 0-d field for restarts, providing the metadata as individual arguments.
- Parameters
[in] f_ptr: A pointer to the field to be read or written[in] name: variable name to be used in the restart file[in] mandatory: If true, the run will abort if this field is not successfully read from the restart file.cs: A pointer to a MOM_restart_CS object (intent in/out)[in] longname: variable long name[in] units: variable units[in] t_grid: time description: s, p, or 1, ‘s’ if absent
-
logical function mom_restart::query_initialized_name(name name, CS CS) query_initialized_name determines whether a named field has been successfully read from a restart file yet.
- Parameters
[in] name: The name of the field that is being queriedcs: A pointer to a MOM_restart_CS object (intent in)
-
logical function mom_restart::query_initialized_0d(f_ptr f_ptr, CS CS) Indicate whether the field pointed to by f_ptr has been initialized from a restart file.
- Parameters
[in] f_ptr: A pointer to the field that is being queriedcs: A pointer to a MOM_restart_CS object (intent in)
-
logical function mom_restart::query_initialized_1d(f_ptr f_ptr, CS CS) Indicate whether the field pointed to by f_ptr has been initialized from a restart file.
- Parameters
[in] f_ptr: A pointer to the field that is being queriedcs: A pointer to a MOM_restart_CS object (intent in)
-
logical function mom_restart::query_initialized_2d(f_ptr f_ptr, CS CS) Indicate whether the field pointed to by f_ptr has been initialized from a restart file.
- Parameters
[in] f_ptr: A pointer to the field that is being queriedcs: A pointer to a MOM_restart_CS object (intent in)
-
logical function mom_restart::query_initialized_3d(f_ptr f_ptr, CS CS) Indicate whether the field pointed to by f_ptr has been initialized from a restart file.
- Parameters
[in] f_ptr: A pointer to the field that is being queriedcs: A pointer to a MOM_restart_CS object (intent in)
-
logical function mom_restart::query_initialized_4d(f_ptr f_ptr, CS CS) Indicate whether the field pointed to by f_ptr has been initialized from a restart file.
- Parameters
[in] f_ptr: A pointer to the field that is being queriedcs: A pointer to a MOM_restart_CS object (intent in)
-
logical function mom_restart::query_initialized_0d_name(f_ptr f_ptr, name name, CS CS) Indicate whether the field pointed to by f_ptr or with the specified variable name has been initialized from a restart file.
- Parameters
[in] f_ptr: A pointer to the field that is being queried[in] name: The name of the field that is being queriedcs: A pointer to a MOM_restart_CS object (intent in)
-
logical function mom_restart::query_initialized_1d_name(f_ptr f_ptr, name name, CS CS) Indicate whether the field pointed to by f_ptr or with the specified variable name has been initialized from a restart file.
- Parameters
[in] f_ptr: A pointer to the field that is being queried[in] name: The name of the field that is being queriedcs: A pointer to a MOM_restart_CS object (intent in)
-
logical function mom_restart::query_initialized_2d_name(f_ptr f_ptr, name name, CS CS) Indicate whether the field pointed to by f_ptr or with the specified variable name has been initialized from a restart file.
- Parameters
[in] f_ptr: A pointer to the field that is being queried[in] name: The name of the field that is being queriedcs: A pointer to a MOM_restart_CS object (intent in)
-
logical function mom_restart::query_initialized_3d_name(f_ptr f_ptr, name name, CS CS) Indicate whether the field pointed to by f_ptr or with the specified variable name has been initialized from a restart file.
- Parameters
[in] f_ptr: A pointer to the field that is being queried[in] name: The name of the field that is being queriedcs: A pointer to a MOM_restart_CS object (intent in)
-
logical function mom_restart::query_initialized_4d_name(f_ptr f_ptr, name name, CS CS) Indicate whether the field pointed to by f_ptr or with the specified variable name has been initialized from a restart file.
- Parameters
[in] f_ptr: A pointer to the field that is being queried[in] name: The name of the field that is being queriedcs: A pointer to a MOM_restart_CS object (intent in)
-
subroutine, public mom_restart::save_restart(directory directory, time time, G G, CS CS, time_stamped time_stamped, filename filename, GV GV) save_restart saves all registered variables to restart files.
- Parameters
[in] directory: The directory where the restart files are to be written[in] time: The current model time[inout] g: The ocean’s grid structurecs: The control structure returned by a previous call to restart_init.[in] time_stamped: If present and true, add time-stamp to the restart file names.[in] filename: A filename that overrides the name in CSrestartfile.[in] gv: The ocean’s vertical grid structure
-
subroutine, public mom_restart::restore_state(filename filename, directory directory, day day, G G, CS CS) restore_state reads the model state from previously generated files. All restart variables are read from the first file in the input filename list in which they are found.
- Parameters
[in] filename: The list of restart file names or a single character ‘r’ to read automatically named files.[in] directory: The directory in which to find restart files[out] day: The time of the restarted run[in] g: The ocean’s grid structurecs: The control structure returned by a previous call to restart_init.
-
logical function, public mom_restart::restart_files_exist(filename filename, directory directory, G G, CS CS) restart_files_exist determines whether any restart files exist.
- Return
The function result, which indicates whether any of the explicitly or automatically named restart files exist in directory.
- Parameters
[in] filename: The list of restart file names or a single character ‘r’ to read automatically named files.[in] directory: The directory in which to find restart files[in] g: The ocean’s grid structurecs: The control structure returned by a previous call to restart_init.
-
logical function, public mom_restart::determine_is_new_run(filename filename, directory directory, G G, CS CS) determine_is_new_run determines from the value of filename and the existence automatically named restart files in directory whether this would be a new, and as a side effect stores this information in CS.
- Return
The function result, which indicates whether this is a new run, based on the value of filename and whether restart files exist.
- Parameters
[in] filename: The list of restart file names or a single character ‘r’ to read automatically named files.[in] directory: The directory in which to find restart files[in] g: The ocean’s grid structurecs: The control structure returned by a previous call to restart_init.
-
logical function, public mom_restart::is_new_run(CS CS) is_new_run returns whether this is going to be a new run based on the information stored in CS by a previous call to determine_is_new_run.
- Return
The function result, which indicates whether this is a new run, based on the value of filename and whether restart files exist.
- Parameters
cs: The control structure returned by a previous call to restart_init.
-
integer function mom_restart::open_restart_units(filename filename, directory directory, G G, CS CS, units units, file_paths file_paths, global_files global_files) open_restart_units determines the number of existing restart files and optionally opens them and returns unit ids, paths and whether the files are global or spatially decomposed.
- Return
The number of files (both automatically named restart files and others explicitly in filename) that have been opened.
- Parameters
[in] filename: The list of restart file names or a single character ‘r’ to read automatically named files.[in] directory: The directory in which to find restart files[in] g: The ocean’s grid structurecs: The control structure returned by a previous call to restart_init.[out] units: The mpp units of all opened files.[out] file_paths: The full paths to open files.[out] global_files: True if a file is global.
-
subroutine, public mom_restart::restart_init(param_file param_file, CS CS, restart_root restart_root) Initialize this module and set up a restart control structure.
- Parameters
[in] param_file: A structure to parse for run-time parameterscs: A pointer to a MOM_restart_CS object that is allocated here[in] restart_root: A filename root that overrides the value
-
subroutine, public mom_restart::restart_init_end(CS CS) Indicate that all variables have now been registered.
- Parameters
cs: A pointer to a MOM_restart_CS object
-
subroutine, public mom_restart::restart_end(CS CS) Deallocate memory associated with a MOM_restart_CS variable.
- Parameters
cs: A pointer to a MOM_restart_CS object
-
subroutine
get_checksum_loop_ranges(G G, pos pos, isL isL, ieL ieL, jsL jsL, jeL jeL)¶ Return bounds for computing checksums to store in restart files.
- Parameters
[in] g: The ocean’s grid structure[in] pos: An integer indicating staggering of variable[out] isl: i-start for checksum[out] iel: i-end for checksum[out] jsl: j-start for checksum[out] jel: j-end for checksum
-
-
namespace
mom_safe_alloc¶ Convenience functions for safely allocating memory without accidentally reallocating pointer and causing memory leaks.
Functions
-
subroutine
safe_alloc_ptr_1d(ptr ptr, i1 i1, i2 i2)¶ Allocate a pointer to a 1-d array.
- Parameters
ptr: A pointer to allocate[in] i1: The size of the array, or its starting index if i2 is present[in] i2: The ending index of the array
-
subroutine
safe_alloc_ptr_2d_2arg(ptr ptr, ni ni, nj nj)¶ Allocate a pointer to a 2-d array based on its dimension sizes.
- Parameters
ptr: A pointer to allocate[in] ni: The size of the 1st dimension of the array[in] nj: The size of the 2nd dimension of the array
-
subroutine
safe_alloc_ptr_3d_3arg(ptr ptr, ni ni, nj nj, nk nk)¶ Allocate a pointer to a 3-d array based on its dimension sizes.
- Parameters
ptr: A pointer to allocate[in] ni: The size of the 1st dimension of the array[in] nj: The size of the 2nd dimension of the array[in] nk: The size of the 3rd dimension of the array
-
subroutine
safe_alloc_ptr_2d(ptr ptr, is is, ie ie, js js, je je)¶ Allocate a pointer to a 2-d array based on its index starting and ending values.
- Parameters
ptr: A pointer to allocate[in] is: The start index to allocate for the 1st dimension[in] ie: The end index to allocate for the 1st dimension[in] js: The start index to allocate for the 2nd dimension[in] je: The end index to allocate for the 2nd dimension
-
subroutine
safe_alloc_ptr_3d(ptr ptr, is is, ie ie, js js, je je, nk nk)¶ Allocate a pointer to a 3-d array based on its index starting and ending values.
- Parameters
ptr: A pointer to allocate[in] is: The start index to allocate for the 1st dimension[in] ie: The end index to allocate for the 1st dimension[in] js: The start index to allocate for the 2nd dimension[in] je: The end index to allocate for the 2nd dimension[in] nk: The size to allocate for the 3rd dimension
-
subroutine
safe_alloc_ptr_3d_6arg(ptr ptr, is is, ie ie, js js, je je, ks ks, ke ke)¶ Allocate a pointer to a 3-d array based on its index starting and ending values.
- Parameters
ptr: A pointer to allocate[in] is: The start index to allocate for the 1st dimension[in] ie: The end index to allocate for the 1st dimension[in] js: The start index to allocate for the 2nd dimension[in] je: The end index to allocate for the 2nd dimension[in] ks: The start index to allocate for the 3rd dimension[in] ke: The end index to allocate for the 3rd dimension
-
subroutine
safe_alloc_allocatable_2d(ptr ptr, is is, ie ie, js js, je je)¶ Allocate a 2-d allocatable array based on its index starting and ending values.
- Parameters
ptr: An allocatable array to allocate[in] is: The start index to allocate for the 1st dimension[in] ie: The end index to allocate for the 1st dimension[in] js: The start index to allocate for the 2nd dimension[in] je: The end index to allocate for the 2nd dimension
-
subroutine
safe_alloc_allocatable_3d(ptr ptr, is is, ie ie, js js, je je, nk nk)¶ Allocate a 3-d allocatable array based on its index starting and ending values and k-index size.
- Parameters
ptr: An allocatable array to allocate[in] is: The start index to allocate for the 1st dimension[in] ie: The end index to allocate for the 1st dimension[in] js: The start index to allocate for the 2nd dimension[in] je: The end index to allocate for the 2nd dimension[in] nk: The size to allocate for the 3rd dimension
-
subroutine
safe_alloc_allocatable_3d_6arg(ptr ptr, is is, ie ie, js js, je je, ks ks, ke ke)¶ Allocate a 3-d allocatable array based on its 6 index starting and ending values.
- Parameters
ptr: An allocatable array to allocate[in] is: The start index to allocate for the 1st dimension[in] ie: The end index to allocate for the 1st dimension[in] js: The start index to allocate for the 2nd dimension[in] je: The end index to allocate for the 2nd dimension[in] ks: The start index to allocate for the 3rd dimension[in] ke: The end index to allocate for the 3rd dimension
-
subroutine
-
namespace
mom_set_diffusivity¶ Calculate vertical diffusivity from all mixing processes.
Unnamed Group
-
integer
id_clock_kappashear¶ CPU time clocks.
-
integer
id_clock_cvmix_ddiff¶ CPU time clocks.
-
subroutine, public mom_set_diffusivity::set_diffusivity(u u, v v, h h, u_h u_h, v_h v_h, tv tv, fluxes fluxes, optics optics, visc visc, dt_in_T dt_in_T, G G, GV GV, US US, CS CS, Kd_lay Kd_lay, Kd_int Kd_int) Sets the interior vertical diffusion of scalars due to the following processes:
Shear-driven mixing: two options, Jackson et at. and KPP interior;
Background mixing via CVMix (Bryan-Lewis profile) or the scheme described by Harrison & Hallberg, JPO 2008;
Double-diffusion, old method and new method via CVMix;
Tidal mixing: many options available, see MOM_tidal_mixing.F90; In addition, this subroutine has the option to set the interior vertical viscosity associated with processes 1,2 and 4 listed above, which is stored in viscKv_slow. Vertical viscosity due to shear-driven mixing is passed via viscKv_shear
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] u: The zonal velocity [m s-1].[in] v: The meridional velocity [m s-1].[in] h: Layer thicknesses [H ~> m or kg m-2].[in] u_h: Zonal velocity interpolated to h points [m s-1].[in] v_h: Meridional velocity interpolated to h points [m s-1].[inout] tv: Structure with pointers to thermodynamic fields. Out is for tvTempxPmE.[in] fluxes: A structure of thermodynamic surface fluxesoptics: A structure describing the optical properties of the ocean.[inout] visc: Structure containing vertical viscosities, bottom boundary layer properies, and related fields.[in] dt_in_t: Time increment [s].cs: Module control structure.[out] kd_lay: Diapycnal diffusivity of each layer [Z2 T-1 ~> m2 s-1].[out] kd_int: Diapycnal diffusivity at each interface [Z2 T-1 ~> m2 s-1].
-
subroutine
find_tke_to_kd(h h, tv tv, dRho_int dRho_int, N2_lay N2_lay, j j, dt dt, G G, GV GV, US US, CS CS, TKE_to_Kd TKE_to_Kd, maxTKE maxTKE, kb kb)¶ Convert turbulent kinetic energy to diffusivity.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[in] h: Layer thicknesses [H ~> m or kg m-2][in] tv: Structure containing pointers to any available thermodynamic fields.[in] drho_int: Change in locally referenced potential density across each interface [kg m-3].[in] n2_lay: The squared buoyancy frequency of the layers [T-2 ~> s-2].[in] j: j-index of row to work on[in] dt: Time increment [T ~> s].cs: Diffusivity control structure[out] tke_to_kd: The conversion rate between the TKE dissipated within a layer and the diapycnal diffusivity witin that layer, usually (~Rho_0 / (G_Earth * dRho_lay)) [Z2 T-1 / Z3 T-3 = T2 Z-1 ~> s2 m-1][out] maxtke: The energy required to for a layer to entrain to its maximum realizable thickness [Z3 T-3 ~> m3 s-3][out] kb: Index of lightest layer denser than the buffer layer, or -1 without a bulk mixed layer.
-
subroutine
find_n2(h h, tv tv, T_f T_f, S_f S_f, fluxes fluxes, j j, G G, GV GV, US US, CS CS, dRho_int dRho_int, N2_lay N2_lay, N2_int N2_int, N2_bot N2_bot)¶ Calculate Brunt-Vaisala frequency, N^2.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[in] h: Layer thicknesses [H ~> m or kg m-2][in] tv: Structure containing pointers to any available thermodynamic fields.[in] t_f: layer temperature with the values in massless layers[in] s_f: Layer salinities with values in massless[in] fluxes: A structure of thermodynamic surface fluxes[in] j: j-index of row to work oncs: Diffusivity control structure[out] drho_int: Change in locally referenced potential density[out] n2_int: The squared buoyancy frequency at the interfaces [T-2 ~> s-2].[out] n2_lay: The squared buoyancy frequency of the layers [T-2 ~> s-2].[out] n2_bot: The near-bottom squared buoyancy frequency [T-2 ~> s-2].
-
subroutine
double_diffusion(tv tv, h h, T_f T_f, S_f S_f, j j, G G, GV GV, US US, CS CS, Kd_T_dd Kd_T_dd, Kd_S_dd Kd_S_dd)¶ This subroutine sets the additional diffusivities of temperature and salinity due to double diffusion, using the same functional form as is used in MOM4.1, and taken from an NCAR technical note (REF?) that updates what was in Large et al. (1994). All the coefficients here should probably be made run-time variables rather than hard-coded constants.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] tv: Structure containing pointers to any available thermodynamic fields; absent fields have NULL ptrs.[in] h: Layer thicknesses [H ~> m or kg m-2].[in] t_f: layer temperatures with the values in massless layers[in] s_f: Layer salinities with values in massless[in] j: Meridional index upon which to work.cs: Module control structure.[out] kd_t_dd: Interface double diffusion diapycnal[out] kd_s_dd: Interface double diffusion diapycnal
-
subroutine
add_drag_diffusivity(h h, u u, v v, tv tv, fluxes fluxes, visc visc, j j, TKE_to_Kd TKE_to_Kd, maxTKE maxTKE, kb kb, G G, GV GV, US US, CS CS, Kd_lay Kd_lay, Kd_int Kd_int, Kd_BBL Kd_BBL)¶ This routine adds diffusion sustained by flow energy extracted by bottom drag.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[in] u: The zonal velocity [m s-1][in] v: The meridional velocity [m s-1][in] h: Layer thicknesses [H ~> m or kg m-2][in] tv: Structure containing pointers to any available thermodynamic fields.[in] fluxes: A structure of thermodynamic surface fluxes[in] visc: Structure containing vertical viscosities, bottom boundary layer properies, and related fields[in] j: j-index of row to work on[in] tke_to_kd: The conversion rate between the TKE TKE dissipated within a layer and the diapycnal diffusivity witin that layer, usually (~Rho_0 / (G_Earth * dRho_lay)) [Z2 T-1 / Z3 T-3 = T2 Z-1 ~> s2 m-1][in] maxtke: The energy required to for a layer to entrain to its maximum-realizable thickness [m3 T-3 ~> m3 s-3][in] kb: Index of lightest layer denser than the buffer layer, or -1 without a bulk mixed layercs: Diffusivity control structure[inout] kd_lay: The diapycnal diffusvity in layers,[inout] kd_int: The diapycnal diffusvity at interfaces,kd_bbl: Interface BBL diffusivity [Z2 T-1 ~> m2 s-1].
-
subroutine
add_lotw_bbl_diffusivity(h h, u u, v v, tv tv, fluxes fluxes, visc visc, j j, N2_int N2_int, G G, GV GV, US US, CS CS, Kd_lay Kd_lay, Kd_int Kd_int, Kd_BBL Kd_BBL)¶ Calculates a BBL diffusivity use a Prandtl number 1 diffusivitiy with a law of the wall turbulent viscosity, up to a BBL height where the energy used for mixing has consumed the mechanical TKE input.
- Parameters
[in] g: Grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[in] u: u component of flow [m s-1][in] v: v component of flow [m s-1][in] h: Layer thickness [H ~> m or kg m-2][in] tv: Structure containing pointers to any available thermodynamic fields.[in] fluxes: Surface fluxes structure[in] visc: Structure containing vertical viscosities, bottom boundary layer properies, and related fields.[in] j: j-index of row to work on[in] n2_int: Square of Brunt-Vaisala at interfaces [T-2 ~> s-2]cs: Diffusivity control structure[inout] kd_lay: Layer net diffusivity [Z2 T-1 ~> m2 s-1][inout] kd_int: Interface net diffusivity [Z2 T-1 ~> m2 s-1]kd_bbl: Interface BBL diffusivity [Z2 T-1 ~> m2 s-1]
-
subroutine
add_mlrad_diffusivity(h h, fluxes fluxes, j j, G G, GV GV, US US, CS CS, Kd_lay Kd_lay, TKE_to_Kd TKE_to_Kd, Kd_int Kd_int)¶ This routine adds effects of mixed layer radiation to the layer diffusivities.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[in] h: Layer thicknesses [H ~> m or kg m-2][in] fluxes: Surface fluxes structurecs: Diffusivity control structure[inout] kd_lay: The diapycnal diffusvity in layers [Z2 T-1 ~> m2 s-1].[in] j: The j-index to work on[in] tke_to_kd: The conversion rate between the TKE TKE dissipated within a layer and the diapycnal diffusivity witin that layer, usually (~Rho_0 / (G_Earth * dRho_lay)) [Z2 T-1 / Z3 T-3 = T2 Z-1 ~> s2 m-1][inout] kd_int: The diapycnal diffusvity at interfaces
-
subroutine, public mom_set_diffusivity::set_bbl_tke(u u, v v, h h, fluxes fluxes, visc visc, G G, GV GV, US US, CS CS) This subroutine calculates several properties related to bottom boundary layer turbulence.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[in] u: The zonal velocity [m s-1][in] v: The meridional velocity [m s-1][in] h: Layer thicknesses [H ~> m or kg m-2][in] fluxes: A structure of thermodynamic surface fluxes[in] visc: Structure containing vertical viscosities, bottom boundary layer properies, and related fields.cs: Diffusivity control structure
-
subroutine
set_density_ratios(h h, tv tv, kb kb, G G, GV GV, US US, CS CS, j j, ds_dsp1 ds_dsp1, rho_0 rho_0)¶ - Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] h: Layer thicknesses [H ~> m or kg m-2].[in] tv: Structure containing pointers to any available thermodynamic fields; absent fields have NULL ptrs.[in] kb: Index of lightest layer denser than the buffer layer, or -1 without a bulk mixed layer.[in] us: A dimensional unit scaling typecs: Control structure returned by previous call to diabatic_entrain_init.[in] j: Meridional index upon which to work.[out] ds_dsp1: Coordinate variable (sigma-2) difference across an interface divided by the difference across the interface below it [nondim][in] rho_0: Layer potential densities relative to
-
subroutine, public mom_set_diffusivity::set_diffusivity_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS, int_tide_CSp int_tide_CSp, tm_CSp tm_CSp, halo_TS halo_TS) - Parameters
[in] time: The current model time[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters.[inout] diag: A structure used to regulate diagnostic output.cs: pointer set to point to the module control structure.int_tide_csp: pointer to the internal tides control structure (BDM)tm_csp: pointer to tidal mixing control structure[out] halo_ts: The halo size of tracer points that must be valid for the calculations in set_diffusivity.
-
subroutine, public mom_set_diffusivity::set_diffusivity_end(CS CS) Clear pointers and dealocate memory.
- Parameters
cs: Control structure for this module
-
integer
-
namespace
mom_set_visc¶ Calculates various values related to the bottom boundary layer, such as the viscosity and thickness of the BBL (set_viscous_BBL).
This would also be the module in which other viscous quantities that are flow-independent might be set. This information is transmitted to other modules via a vertvisc type structure.
The same code is used for the two velocity components, by indirectly referencing the velocities and defining a handful of direction-specific defined variables.
Unnamed Group
-
subroutine, public mom_set_visc::set_viscous_bbl(u u, v v, h h, tv tv, visc visc, G G, GV GV, US US, CS CS, symmetrize symmetrize) Calculates the thickness of the bottom boundary layer and the viscosity within that layer. A drag law is used, either linearized about an assumed bottom velocity or using the actual near-bottom velocities combined with an assumed unresolved velocity. The bottom boundary layer thickness is limited by a combination of stratification and rotation, as in the paper of Killworth and Edwards, JPO 1999. It is not necessary to calculate the thickness and viscosity every time step; instead previous values may be used.
- Parameters
[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] u: The zonal velocity [m s-1].[in] v: The meridional velocity [m s-1].[in] h: Layer thicknesses [H ~> m or kg m-2].[in] tv: A structure containing pointers to any available thermodynamic fields. Absent fields have NULL ptrs..[inout] visc: A structure containing vertical viscosities and related fields.cs: The control structure returned by a previous call to vertvisc_init.[in] symmetrize: If present and true, do extra calculations of those values in visc that would be calculated with symmetric memory.
-
real function mom_set_visc::set_v_at_u(v v, h h, G G, i i, j j, k k, mask2dCv mask2dCv, OBC OBC) This subroutine finds a thickness-weighted value of v at the u-points.
- Return
The retur value of v at u points [m s-1].
- Parameters
[in] g: The ocean’s grid structure[in] v: The meridional velocity [m s-1][in] h: Layer thicknesses [H ~> m or kg m-2][in] i: The i-index of the u-location to work on.[in] j: The j-index of the u-location to work on.[in] k: The k-index of the u-location to work on.[in] mask2dcv: A multiplicative mask of the v-pointsobc: A pointer to an open boundary condition structure
-
real function mom_set_visc::set_u_at_v(u u, h h, G G, i i, j j, k k, mask2dCu mask2dCu, OBC OBC) This subroutine finds a thickness-weighted value of u at the v-points.
- Return
The return value of u at v points [m s-1].
- Parameters
[in] g: The ocean’s grid structure[in] u: The zonal velocity [m s-1][in] h: Layer thicknesses [H ~> m or kg m-2][in] i: The i-index of the u-location to work on.[in] j: The j-index of the u-location to work on.[in] k: The k-index of the u-location to work on.[in] mask2dcu: A multiplicative mask of the u-pointsobc: A pointer to an open boundary condition structure
-
subroutine, public mom_set_visc::set_viscous_ml(u u, v v, h h, tv tv, forces forces, visc visc, dt dt, G G, GV GV, US US, CS CS, symmetrize symmetrize) Calculates the thickness of the surface boundary layer for applying an elevated viscosity.
A bulk Richardson criterion or the thickness of the topmost NKML layers (with a bulk mixed layer) are currently used. The thicknesses are given in terms of fractional layers, so that this thickness will move as the thickness of the topmost layers change.
- Parameters
[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] u: The zonal velocity [m s-1].[in] v: The meridional velocity [m s-1].[in] h: Layer thicknesses [H ~> m or kg m-2].[in] tv: A structure containing pointers to any available thermodynamic fields. Absent fields have NULL ptrs.[in] forces: A structure with the driving mechanical forces[inout] visc: A structure containing vertical viscosities and related fields.[in] dt: Time increment [s].cs: The control structure returned by a previous call to vertvisc_init.[in] symmetrize: If present and true, do extra calculations of those values in visc that would be calculated with symmetric memory.
-
subroutine, public mom_set_visc::set_visc_register_restarts(HI HI, GV GV, param_file param_file, visc visc, restart_CS restart_CS) Register any fields associated with the vertvisc_type.
- Parameters
[in] hi: A horizontal index type structure.[in] gv: The ocean’s vertical grid structure.[in] param_file: A structure to parse for run-time parameters.[inout] visc: A structure containing vertical viscosities and related fields. Allocated here.restart_cs: A pointer to the restart control structure.
-
subroutine, public mom_set_visc::set_visc_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, visc visc, CS CS, restart_CS restart_CS, OBC OBC) Initializes the MOM_set_visc control structure.
- Parameters
[in] time: The current model time.[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters.[inout] diag: A structure that is used to regulate diagnostic output.[inout] visc: A structure containing vertical viscosities and related fields. Allocated here.cs: A pointer that is set to point to the control structure for this modulerestart_cs: A pointer to the restart control structure.obc: A pointer to an open boundary condition structure
-
subroutine, public mom_set_visc::set_visc_end(visc visc, CS CS) This subroutine dellocates any memory in the set_visc control structure.
- Parameters
[inout] visc: A structure containing vertical viscosities and related fields. Elements are deallocated here.cs: The control structure returned by a previous call to vertvisc_init.
-
Code that initializes fixed aspects of the model grid, such as horizontal grid metrics, topography and Coriolis, and can be shared between components.
Functions
-
subroutine, public mom_shared_initialization::mom_shared_init_init(PF PF) MOM_shared_init_init just writes the code version.
- Parameters
[in] pf: A structure indicating the open file to parse for model parameter values.
-
subroutine, public mom_shared_initialization::mom_initialize_rotation(f f, G G, PF PF, US US) MOM_initialize_rotation makes the appropriate call to set up the Coriolis parameter.
- Parameters
[in] g: The dynamic horizontal grid type[out] f: The Coriolis parameter [T-1 ~> s-1][in] pf: Parameter file structure[in] us: A dimensional unit scaling type
-
subroutine, public mom_shared_initialization::mom_calculate_grad_coriolis(dF_dx dF_dx, dF_dy dF_dy, G G, US US) Calculates the components of grad f (Coriolis parameter)
- Parameters
[inout] g: The dynamic horizontal grid type[out] df_dx: x-component of grad f [T-1 m-1 ~> s-1 m-1][out] df_dy: y-component of grad f [T-1 m-1 ~> s-1 m-1][in] us: A dimensional unit scaling type
-
real function, public mom_shared_initialization::diagnosemaximumdepth(D D, G G) Return the global maximum ocean bottom depth in the same units as the input depth.
- Return
The global maximum ocean bottom depth in m or Z
- Parameters
[in] g: The dynamic horizontal grid type[in] d: Ocean bottom depth in m or Z
-
subroutine, public mom_shared_initialization::initialize_topography_from_file(D D, G G, param_file param_file, US US) Read gridded depths from file.
- Parameters
[in] g: The dynamic horizontal grid type[out] d: Ocean bottom depth in m or Z if US is present[in] param_file: Parameter file structure[in] us: A dimensional unit scaling type
-
subroutine, public mom_shared_initialization::apply_topography_edits_from_file(D D, G G, param_file param_file, US US) Applies a list of topography overrides read from a netcdf file.
- Parameters
[in] g: The dynamic horizontal grid type[inout] d: Ocean bottom depth in m or Z if US is present[in] param_file: Parameter file structure[in] us: A dimensional unit scaling type
-
subroutine, public mom_shared_initialization::initialize_topography_named(D D, G G, param_file param_file, topog_config topog_config, max_depth max_depth, US US) initialize the bathymetry based on one of several named idealized configurations
- Parameters
[in] g: The dynamic horizontal grid type[out] d: Ocean bottom depth in m or Z if US is present[in] param_file: Parameter file structure[in] topog_config: The name of an idealized topographic configuration[in] max_depth: Maximum depth of model in the units of D[in] us: A dimensional unit scaling type
-
subroutine, public mom_shared_initialization::limit_topography(D D, G G, param_file param_file, max_depth max_depth, US US) limit_topography ensures that min_depth < D(x,y) < max_depth
- Parameters
[in] g: The dynamic horizontal grid type[inout] d: Ocean bottom depth in m or Z if US is present[in] param_file: Parameter file structure[in] max_depth: Maximum depth of model in the units of D[in] us: A dimensional unit scaling type
-
subroutine, public mom_shared_initialization::set_rotation_planetary(f f, G G, param_file param_file, US US) This subroutine sets up the Coriolis parameter for a sphere.
- Parameters
[in] g: The dynamic horizontal grid[out] f: Coriolis parameter (vertical component) [T-1 ~> s-1][in] param_file: A structure to parse for run-time parameters[in] us: A dimensional unit scaling type
-
subroutine, public mom_shared_initialization::set_rotation_beta_plane(f f, G G, param_file param_file, US US) This subroutine sets up the Coriolis parameter for a beta-plane or f-plane.
- Parameters
[in] g: The dynamic horizontal grid[out] f: Coriolis parameter (vertical component) [T-1 ~> s-1][in] param_file: A structure to parse for run-time parameters[in] us: A dimensional unit scaling type
-
subroutine, public mom_shared_initialization::initialize_grid_rotation_angle(G G, PF PF) initialize_grid_rotation_angle initializes the arrays with the sine and cosine of the angle between logical north on the grid and true north.
- Parameters
[inout] g: The dynamic horizontal grid[in] pf: A structure indicating the open file to parse for model parameter values.
-
real function mom_shared_initialization::modulo_around_point(x x, xc xc, Lx Lx) Return the modulo value of x in an interval [xc-(Lx/2) xc+(Lx/2)] If Lx<=0, then it returns x without applying modulo arithmetic.
- Return
x shifted by an integer multiple of Lx to be close to xc.
- Parameters
[in] x: Value to which to apply modulo arithmetic[in] xc: Center of modulo range[in] lx: Modulo range width
-
subroutine, public mom_shared_initialization::reset_face_lengths_named(G G, param_file param_file, name name, US US) This subroutine sets the open face lengths at selected points to restrict passages to their observed widths based on a named set of sizes.
- Parameters
[inout] g: The dynamic horizontal grid[in] param_file: A structure to parse for run-time parameters[in] name: The name for the set of face lengths. Only “global_1deg” is currently implemented.[in] us: A dimensional unit scaling type
-
subroutine, public mom_shared_initialization::reset_face_lengths_file(G G, param_file param_file, US US) This subroutine sets the open face lengths at selected points to restrict passages to their observed widths from a arrays read from a file.
- Parameters
[inout] g: The dynamic horizontal grid[in] param_file: A structure to parse for run-time parameters[in] us: A dimensional unit scaling type
-
subroutine, public mom_shared_initialization::reset_face_lengths_list(G G, param_file param_file, US US) This subroutine sets the open face lengths at selected points to restrict passages to their observed widths from a list read from a file.
- Parameters
[inout] g: The dynamic horizontal grid[in] param_file: A structure to parse for run-time parameters[in] us: A dimensional unit scaling type
-
subroutine, public mom_shared_initialization::read_face_length_list(iounit iounit, filename filename, num_lines num_lines, lines lines) This subroutine reads and counts the non-blank lines in the face length list file, after removing comments.
- Parameters
[in] iounit: An open I/O unit number for the file[in] filename: The name of the face-length file to read[out] num_lines: The number of non-blank lines in the filelines: The non-blank lines, after removing comments
-
subroutine, public mom_shared_initialization::set_velocity_depth_max(G G) Set the bathymetry at velocity points to be the maximum of the depths at the neighoring tracer points.
- Parameters
[inout] g: The dynamic horizontal grid
-
subroutine, public mom_shared_initialization::set_velocity_depth_min(G G) Set the bathymetry at velocity points to be the minimum of the depths at the neighoring tracer points.
- Parameters
[inout] g: The dynamic horizontal grid
-
subroutine, public mom_shared_initialization::compute_global_grid_integrals(G G) Pre-compute global integrals of grid quantities (like masked ocean area) for later use in reporting diagnostics.
- Parameters
[inout] g: The dynamic horizontal grid
-
subroutine, public mom_shared_initialization::write_ocean_geometry_file(G G, param_file param_file, directory directory, geom_file geom_file, US US) Write out a file describing the topography, Coriolis parameter, grid locations and various other fixed fields from the grid.
- Parameters
[inout] g: The dynamic horizontal grid[in] param_file: Parameter file structure[in] directory: The directory into which to place the geometry file.[in] geom_file: If present, the name of the geometry file (otherwise the file is “ocean_geometry”)[in] us: A dimensional unit scaling type
-
-
namespace
mom_spatial_means¶ Functions and routines to take area, volume, mass-weighted, layerwise, zonal or meridional means.
Functions
-
real function, public mom_spatial_means::global_area_mean(var var, G G) Return the global area mean of a variable. This uses reproducing sums.
- Parameters
[in] g: The ocean’s grid structure[in] var: The variable to average
-
real function, public mom_spatial_means::global_area_integral(var var, G G) Return the global area integral of a variable. This uses reproducing sums.
- Parameters
[in] g: The ocean’s grid structure[in] var: The variable to integrate
-
real function, dimension( gv %ke), public mom_spatial_means::global_layer_mean(var var, h h, G G, GV GV) Return the layerwise global thickness-weighted mean of a variable. This uses reproducing sums.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] var: The variable to average[in] h: Layer thicknesses [H ~> m or kg m-2]
-
real function, public mom_spatial_means::global_volume_mean(var var, h h, G G, GV GV) Find the global thickness-weighted mean of a variable. This uses reproducing sums.
- Return
The thickness-weighted average of var
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] var: The variable being averaged[in] h: Layer thicknesses [H ~> m or kg m-2]
-
real function, public mom_spatial_means::global_mass_integral(h h, G G, GV GV, var var, on_PE_only on_PE_only) Find the global mass-weighted integral of a variable. This uses reproducing sums.
- Return
The mass-weighted integral of var (or 1) in kg times the units of var
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][in] var: The variable being integrated[in] on_pe_only: If present and true, the sum is only done on the local PE, and it is not order invariant.
-
subroutine, public mom_spatial_means::global_i_mean(array array, i_mean i_mean, G G, mask mask) Determine the global mean of a field along rows of constant i, returning it in a 1-d array using the local indexing. This uses reproducing sums.
- Parameters
[inout] g: The ocean’s grid structure[in] array: The variable being averaged[out] i_mean: Global mean of array along its i-axis[in] mask: An array used for weighting the i-mean
-
subroutine, public mom_spatial_means::global_j_mean(array array, j_mean j_mean, G G, mask mask) Determine the global mean of a field along rows of constant j, returning it in a 1-d array using the local indexing. This uses reproducing sums.
- Parameters
[inout] g: The ocean’s grid structure[in] array: The variable being averaged[out] j_mean: Global mean of array along its j-axis[in] mask: An array used for weighting the j-mean
-
subroutine, public mom_spatial_means::adjust_area_mean_to_zero(array array, G G, scaling scaling) Adjust 2d array such that area mean is zero without moving the zero contour.
- Parameters
[in] g: Grid structure[inout] array: 2D array to be adjusted[out] scaling: The scaling factor used
-
-
namespace
mom_sponge¶ Implements sponge regions in isopycnal mode.
By Robert Hallberg, March 1999-June 2000
This program contains the subroutines that implement sponge regions, in which the stratification and water mass properties are damped toward some profiles. There are three externally callable subroutines in this file.
initialize_sponge determines the mapping from the model variables into the arrays of damped columns. This remapping is done for efficiency and to conserve memory. Only columns which have positive inverse damping times and which are deeper than a supplied depth are placed in sponges. The inverse damping time is also stored in this subroutine, and memory is allocated for all of the reference profiles which will subsequently be provided through calls to set_up_sponge_field. The first two arguments are a two-dimensional array containing the damping rates, and the interface heights to damp towards.
set_up_sponge_field is called to provide a reference profile and the location of the field that will be damped back toward that reference profile. A third argument, the number of layers in the field is also provided, but this should always be nz.
Apply_sponge damps all of the fields that have been registered with set_up_sponge_field toward their reference profiles. The four arguments are the thickness to be damped, the amount of time over which the damping occurs, and arrays to which the movement of fluid into a layer from above and below will be added. The effect on momentum of the sponge may be accounted for later using the movement of water recorded in these later arrays.
Functions
-
subroutine, public mom_sponge::initialize_sponge(Iresttime Iresttime, int_height int_height, G G, param_file param_file, CS CS, GV GV, Iresttime_i_mean Iresttime_i_mean, int_height_i_mean int_height_i_mean) This subroutine determines the number of points which are within sponges in this computational domain. Only points that have positive values of Iresttime and which mask2dT indicates are ocean points are included in the sponges. It also stores the target interface heights.
- Parameters
[in] g: The ocean’s grid structure[in] iresttime: The inverse of the restoring time [s-1].[in] int_height: The interface heights to damp back toward [Z ~> m].[in] param_file: A structure to parse for run-time parameterscs: A pointer that is set to point to the control structure for this module[in] gv: The ocean’s vertical grid structure[in] iresttime_i_mean: The inverse of the restoring time for[in] int_height_i_mean: The interface heights toward which to
-
subroutine, public mom_sponge::init_sponge_diags(Time Time, G G, diag diag, CS CS) This subroutine sets up diagnostics for the sponges. It is separate from initialize_sponge because it requires fields that are not readily available where initialize_sponge is called.
- Parameters
[in] time: The current model time[in] g: The ocean’s grid structure[inout] diag: A structure that is used to regulate diagnostic outputcs: A pointer to the control structure for this module that is set by a previous call to initialize_sponge.
-
subroutine, public mom_sponge::set_up_sponge_field(sp_val sp_val, f_ptr f_ptr, G G, nlay nlay, CS CS, sp_val_i_mean sp_val_i_mean) This subroutine stores the reference profile for the variable whose address is given by f_ptr. nlay is the number of layers in this variable.
- Parameters
[in] g: The ocean’s grid structure[in] sp_val: The reference profiles of the quantity being registered.[in] f_ptr: a pointer to the field which will be damped[in] nlay: the number of layers in this quantitycs: A pointer to the control structure for this module that is set by a previous call to initialize_sponge.[in] sp_val_i_mean: The i-mean reference value for
-
subroutine, public mom_sponge::set_up_sponge_ml_density(sp_val sp_val, G G, CS CS, sp_val_i_mean sp_val_i_mean) This subroutine stores the reference value for mixed layer density. It is handled differently from other values because it is only used in determining which layers can be inflated.
- Parameters
[in] g: The ocean’s grid structure[in] sp_val: The reference values of the mixed layer density [kg m-3]cs: A pointer to the control structure for this module that is set by a previous call to initialize_sponge.[in] sp_val_i_mean: the reference values of the zonal mean mixed
-
subroutine, public mom_sponge::apply_sponge(h h, dt dt, G G, GV GV, ea ea, eb eb, CS CS, Rcv_ml Rcv_ml) This subroutine applies damping to the layers thicknesses, mixed layer buoyancy, and a variety of tracers for every column where there is damping.
- Parameters
[inout] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[inout] h: Layer thicknesses [H ~> m or kg m-2][in] dt: The amount of time covered by this call [s].[inout] ea: An array to which the amount of fluid entrained[inout] eb: An array to which the amount of fluid entrainedcs: A pointer to the control structure for this module that is set by a previous call to initialize_sponge.[inout] rcv_ml: The coordinate density of the mixed layer [kg m-3].
-
subroutine, public mom_sponge::sponge_end(CS CS) This call deallocates any memory in the sponge control structure.
- Parameters
cs: A pointer to the control structure for this module that is set by a previous call to initialize_sponge.
-
-
namespace
mom_state_initialization¶ Initialization functions for state variables, u, v, h, T and S.
Functions
-
subroutine, public mom_state_initialization::mom_initialize_state(u u, v v, h h, tv tv, Time Time, G G, GV GV, US US, PF PF, dirs dirs, restart_CS restart_CS, ALE_CSp ALE_CSp, tracer_Reg tracer_Reg, sponge_CSp sponge_CSp, ALE_sponge_CSp ALE_sponge_CSp, OBC OBC, Time_in Time_in) Initialize temporally evolving fields, either as initial conditions or by reading them from a restart (or saves) file.
- Parameters
[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[out] u: The zonal velocity that is being[out] v: The meridional velocity that is being[out] h: Layer thicknesses [H ~> m or kg m-2][inout] tv: A structure pointing to various thermodynamic variables[inout] time: Time at the start of the run segment.[in] pf: A structure indicating the open file to parse for model parameter values.[in] dirs: A structure containing several relevant directory paths.restart_cs: A pointer to the restart control structure.ale_csp: The ALE control structure for remappingtracer_reg: A pointer to the tracer registrysponge_csp: The layerwise sponge control structure.ale_sponge_csp: The ALE sponge control structure.obc: The open boundary condition control structure.[in] time_in: Time at the start of the run segment. Time_in overrides any value set for Time.
-
subroutine
initialize_thickness_from_file(h h, G G, GV GV, US US, param_file param_file, file_has_thickness file_has_thickness, just_read_params just_read_params)¶ Reads the layer thicknesses or interface heights from a file.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[out] h: The thickness that is being initialized [H ~> m or kg m-2].[in] param_file: A structure indicating the open file to parse for model parameter values.[in] file_has_thickness: If true, this file contains layer thicknesses; otherwise it contains interface heights.[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine
adjustetatofitbathymetry(G G, GV GV, US US, eta eta, h h)¶ Adjust interface heights to fit the bathymetry and diagnose layer thickness.
If the bottom most interface is below the topography then the bottom-most layers are contracted to GVAngstrom_m. If the bottom most interface is above the topography then the entire column is dilated (expanded) to fill the void.
- Remark
{There is a (hard-wired) “tolerance” parameter such that the criteria for adjustment must equal or exceed 10cm.}
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[inout] eta: Interface heights [Z ~> m].[inout] h: Layer thicknesses [H ~> m or kg m-2]
-
subroutine
initialize_thickness_uniform(h h, G G, GV GV, param_file param_file, just_read_params just_read_params)¶ Initializes thickness to be uniform.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[out] h: The thickness that is being initialized [H ~> m or kg m-2].[in] param_file: A structure indicating the open file to parse for model parameter values.[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine
initialize_thickness_list(h h, G G, GV GV, US US, param_file param_file, just_read_params just_read_params)¶ Initialize thickness from a 1D list.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[out] h: The thickness that is being initialized [H ~> m or kg m-2].[in] param_file: A structure indicating the open file to parse for model parameter values.[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine
initialize_thickness_search()¶ Search density space for location of layers (not implemented!)
-
subroutine
convert_thickness(h h, G G, GV GV, US US, tv tv)¶ Converts thickness from geometric to pressure units.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[inout] h: Input geometric layer thicknesses being converted[in] tv: A structure pointing to various thermodynamic variables
-
subroutine
depress_surface(h h, G G, GV GV, US US, param_file param_file, tv tv, just_read_params just_read_params)¶ Depress the sea-surface based on an initial condition file.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[inout] h: Layer thicknesses [H ~> m or kg m-2][in] param_file: A structure to parse for run-time parameters[in] tv: A structure pointing to various thermodynamic variables[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine
trim_for_ice(PF PF, G G, GV GV, US US, ALE_CSp ALE_CSp, tv tv, h h, just_read_params just_read_params)¶ Adjust the layer thicknesses by cutting away the top of each model column at the depth where the hydrostatic pressure matches an imposed surface pressure read from file.
- Parameters
[in] pf: Parameter file structure[in] g: Ocean grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling typeale_csp: ALE control structure[inout] tv: Thermodynamics structure[inout] h: Layer thickness [H ~> m or kg m-2][in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine
cut_off_column_top(nk nk, tv tv, GV GV, G_earth G_earth, depth depth, min_thickness min_thickness, T T, T_t T_t, T_b T_b, S S, S_t S_t, S_b S_b, p_surf p_surf, h h, remap_CS remap_CS, z_tol z_tol)¶ Adjust the layer thicknesses by removing the top of the water column above the depth where the hydrostatic pressure matches p_surf.
- Parameters
[in] nk: Number of layers[in] tv: Thermodynamics structure[in] gv: The ocean’s vertical grid structure.[in] g_earth: Gravitational acceleration [m2 Z-1 s-2 ~> m s-2][in] depth: Depth of ocean column [Z ~> m].[in] min_thickness: Smallest thickness allowed [Z ~> m].[inout] t: Layer mean temperature [degC][in] t_t: Temperature at top of layer [degC][in] t_b: Temperature at bottom of layer [degC][inout] s: Layer mean salinity [ppt][in] s_t: Salinity at top of layer [ppt][in] s_b: Salinity at bottom of layer [ppt][in] p_surf: Imposed pressure on ocean at surface [Pa][inout] h: Layer thickness [H ~> m or kg m-2]remap_cs: Remapping structure for remapping T and S, if associated[in] z_tol: The tolerance with which to find the depth matching the specified pressure [Z ~> m].
-
subroutine
initialize_velocity_from_file(u u, v v, G G, param_file param_file, just_read_params just_read_params)¶ Initialize horizontal velocity components from file.
- Parameters
[in] g: The ocean’s grid structure[out] u: The zonal velocity that is being initialized [m s-1][out] v: The meridional velocity that is being initialized [m s-1][in] param_file: A structure indicating the open file to parse for modelparameter values.[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine
initialize_velocity_zero(u u, v v, G G, param_file param_file, just_read_params just_read_params)¶ Initialize horizontal velocity components to zero.
- Parameters
[in] g: The ocean’s grid structure[out] u: The zonal velocity that is being initialized [m s-1][out] v: The meridional velocity that is being initialized [m s-1][in] param_file: A structure indicating the open file to parse for modelparameter values.[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine
initialize_velocity_uniform(u u, v v, G G, param_file param_file, just_read_params just_read_params)¶ Sets the initial velocity components to uniform.
- Parameters
[in] g: The ocean’s grid structure[out] u: The zonal velocity that is being initialized [m s-1][out] v: The meridional velocity that is being initialized [m s-1][in] param_file: A structure indicating the open file to parse for modelparameter values.[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine
initialize_velocity_circular(u u, v v, G G, param_file param_file, just_read_params just_read_params)¶ Sets the initial velocity components to be circular with no flow at edges of domain and center.
- Parameters
[in] g: The ocean’s grid structure[out] u: The zonal velocity that is being initialized [m s-1][out] v: The meridional velocity that is being initialized [m s-1][in] param_file: A structure indicating the open file to parse for model parameter values.[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine
initialize_temp_salt_from_file(T T, S S, G G, param_file param_file, just_read_params just_read_params)¶ Initializes temperature and salinity from file.
- Parameters
[in] g: The ocean’s grid structure[out] t: The potential temperature that is being initialized [degC][out] s: The salinity that is being initialized [ppt][in] param_file: A structure to parse for run-time parameters[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine
initialize_temp_salt_from_profile(T T, S S, G G, param_file param_file, just_read_params just_read_params)¶ Initializes temperature and salinity from a 1D profile.
- Parameters
[in] g: The ocean’s grid structure[out] t: The potential temperature that is being initialized [degC][out] s: The salinity that is being initialized [ppt][in] param_file: A structure to parse for run-time parameters[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine
initialize_temp_salt_fit(T T, S S, G G, GV GV, param_file param_file, eqn_of_state eqn_of_state, P_Ref P_Ref, just_read_params just_read_params)¶ Initializes temperature and salinity by fitting to density.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[out] t: The potential temperature that is being initialized [degC].[out] s: The salinity that is being initialized [ppt].[in] param_file: A structure to parse for run-time parameters.eqn_of_state: Integer that selects the equatio of state.[in] p_ref: The coordinate-density reference pressure [Pa].[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine
initialize_temp_salt_linear(T T, S S, G G, param_file param_file, just_read_params just_read_params)¶ Initializes T and S with linear profiles according to reference surface layer salinity and temperature and a specified range.
- Remark
Note that the linear distribution is set up with respect to the layer number, not the physical position).
- Parameters
[in] g: The ocean’s grid structure[out] t: The potential temperature that is being initialized [degC][out] s: The salinity that is being initialized [ppt][in] param_file: A structure to parse for run-time parameters[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine
initialize_sponges_file(G G, GV GV, US US, use_temperature use_temperature, tv tv, param_file param_file, CSp CSp, ALE_CSp ALE_CSp, Time Time)¶ This subroutine sets the inverse restoration time (Idamp), and the values towards which the interface heights and an arbitrary number of tracers should be restored within each sponge. The interface height is always subject to damping, and must always be the first registered field.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] use_temperature: If true, T & S are state variables.[in] tv: A structure pointing to various thermodynamic variables.[in] param_file: A structure to parse for run-time parameters.csp: A pointer that is set to point to the control structure for this module (in layered mode).ale_csp: A pointer that is set to point to the control structure for this module (in ALE mode).[in] time: Time at the start of the run segment. Time_in overrides any value set for Time.
-
subroutine
set_velocity_depth_max(G G)¶ This subroutine sets the 4 bottom depths at velocity points to be the maximum of the adjacent depths.
- Parameters
[inout] g: The ocean’s grid structure
-
subroutine
compute_global_grid_integrals(G G)¶ Subroutine to pre-compute global integrals of grid quantities for later use in reporting diagnostics.
- Parameters
[inout] g: The ocean’s grid structure
-
subroutine
set_velocity_depth_min(G G)¶ This subroutine sets the 4 bottom depths at velocity points to be the minimum of the adjacent depths.
- Parameters
[inout] g: The ocean’s grid structure
-
subroutine
mom_temp_salt_initialize_from_z(h h, tv tv, G G, GV GV, US US, PF PF, just_read_params just_read_params)¶ This subroutine determines the isopycnal or other coordinate interfaces and layer potential temperatures and salinities directly from a z-space file on a latitude-longitude grid.
- Parameters
[inout] g: The ocean’s grid structure[out] h: Layer thicknesses being initialized [H ~> m or kg m-2][inout] tv: A structure pointing to various thermodynamic variables including temperature and salinity[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[in] pf: A structure indicating the open file to parse for model parameter values.[in] just_read_params: If present and true, this call will only read parameters without changing h.
Variables
-
character(len=40) mom_state_initialization::mdl = "MOM_state_initialization" This module’s name.
-
-
namespace
mom_string_functions¶ Handy functions for manipulating strings.
By Alistair Adcroft and Robert Hallberg, last updated Sept. 2013.
The functions here perform a set of useful manipulations of character strings. Although they are a part of MOM6, the do not require any other MOM software to be useful.
Functions
-
character(len=len(input_string)) function, public mom_string_functions::lowercase(input_string input_string) Return a string in which all uppercase letters have been replaced by their lowercase counterparts.
- Return
The modified output string
- Parameters
[in] input_string: The string to modify
-
character(len=len(input_string)) function, public mom_string_functions::uppercase(input_string input_string) Return a string in which all uppercase letters have been replaced by their lowercase counterparts.
- Return
The modified output string
- Parameters
[in] input_string: The string to modify
-
character(len=19) function, public mom_string_functions::left_int(i i) Returns a character string of a left-formatted integer e.g. “123 ” (assumes 19 digit maximum)
- Return
The output string
- Parameters
[in] i: The integer to convert to a string
-
character(len=1320) function, public mom_string_functions::left_ints(i i) Returns a character string of a comma-separated, compact formatted, integers e.g. “1, 2, 3, 4”.
- Return
The output string
- Parameters
[in] i: The array of integers to convert to a string
-
character(len=32) function, public mom_string_functions::left_real(val val) Returns a left-justified string with a real formatted like ‘(G)’.
- Return
The output string
- Parameters
[in] val: The real variable to convert to a string
-
character(len=1320) function, public mom_string_functions::left_reals(r r, sep sep) Returns a character string of a comma-separated, compact formatted, reals e.g. “1., 2., 5*3., 5.E2”.
- Return
The output string
- Parameters
[in] r: The array of real variables to convert to a string[in] sep: The separator between successive values, by default it is ‘, ‘.
-
logical function mom_string_functions::isformattedfloatequalto(str str, val val) Returns True if the string can be read/parsed to give the exact value of “val”.
- Parameters
[in] str: The string to parse[in] val: The real value to compare with
-
character(len=120) function, public mom_string_functions::extractword(string string, n n) Returns the string corresponding to the nth word in the argument or “” if the string is not long enough. Both spaces and commas are interpreted as separators.
- Parameters
[in] string: The string to scan[in] n: Number of word to extract
-
character(len=120) function, public mom_string_functions::extract_word(string string, separators separators, n n) Returns the string corresponding to the nth word in the argument or “” if the string is not long enough. Words are delineated by the mandatory separators argument.
- Parameters
[in] string: String to scan[in] separators: Characters to use for delineation[in] n: Number of word to extract
-
integer function, public mom_string_functions::extract_integer(string string, separators separators, n n, missing_value missing_value) Returns the integer corresponding to the nth word in the argument.
- Parameters
[in] string: String to scan[in] separators: Characters to use for delineation[in] n: Number of word to extract[in] missing_value: Value to assign if word is missing
-
real function, public mom_string_functions::extract_real(string string, separators separators, n n, missing_value missing_value) Returns the real corresponding to the nth word in the argument.
- Parameters
[in] string: String to scan[in] separators: Characters to use for delineation[in] n: Number of word to extract[in] missing_value: Value to assign if word is missing
-
character(len=120) function, public mom_string_functions::remove_spaces(string string) Returns string with all spaces removed.
- Parameters
[in] string: String to scan
-
logical function, public mom_string_functions::string_functions_unit_tests(verbose verbose) Returns true if a unit test of string_functions fails.
- Parameters
[in] verbose: If true, write results to stdout
-
logical function mom_string_functions::localtests(verbose verbose, str1 str1, str2 str2) True if str1 does not match str2. False otherwise.
- Parameters
[in] verbose: If true, write results to stdout[in] str1: String[in] str2: String
-
logical function mom_string_functions::localtesti(verbose verbose, i1 i1, i2 i2) True if i1 is not equal to i2. False otherwise.
- Parameters
[in] verbose: If true, write results to stdout[in] i1: Integer[in] i2: Integer
-
logical function mom_string_functions::localtestr(verbose verbose, r1 r1, r2 r2) True if r1 is not equal to r2. False otherwise.
- Parameters
[in] verbose: If true, write results to stdout[in] r1: Float[in] r2: Float
-
character(len=len(dir)+2) function, public mom_string_functions::slasher(dir dir) Returns a directory name that is terminated with a “/” or “./” if the argument is an empty string.
- Parameters
[in] dir: A directory to be terminated with a “/” or changed to “./” if it is blank.
-
-
namespace
mom_sum_output¶ Reports integrated quantities for monitoring the model state.
By Robert Hallberg, April 1994 - June 2002
This file contains the subroutine (write_energy) that writes horizontally integrated quantities, such as energies and layer volumes, and other summary information to an output file. Some of these quantities (APE or resting interface height) are defined relative to the global histogram of topography. The subroutine that compiles that histogram (depth_list_setup) is also included in this file.
In addition, if the number of velocity truncations since the previous call to write_energy exceeds maxtrunc or the total energy exceeds a very large threshold, a fatal termination is triggered.
Functions
-
subroutine, public mom_sum_output::mom_sum_output_init(G G, US US, param_file param_file, directory directory, ntrnc ntrnc, Input_start_time Input_start_time, CS CS) MOM_sum_output_init initializes the parameters and settings for the MOM_sum_output module.
- Parameters
[in] g: The ocean’s grid structure.[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters.[in] directory: The directory where the energy file goes.[inout] ntrnc: The integer that stores the number of times the velocity has been truncated since the last call to write_energy.[in] input_start_time: The start time of the simulation.cs: A pointer that is set to point to the control structure for this module.
-
subroutine
mom_sum_output_end(CS CS)¶ MOM_sum_output_end deallocates memory used by the MOM_sum_output module.
- Parameters
cs: The control structure returned by a previous call to MOM_sum_output_init.
-
subroutine, public mom_sum_output::write_energy(u u, v v, h h, tv tv, day day, n n, G G, GV GV, US US, CS CS, tracer_CSp tracer_CSp, OBC OBC, dt_forcing dt_forcing) This subroutine calculates and writes the total model energy, the energy and mass of each layer, and other globally integrated physical quantities.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] u: The zonal velocity [m s-1].[in] v: The meridional velocity [m s-1].[in] h: Layer thicknesses [H ~> m or kg m-2].[in] tv: A structure pointing to various thermodynamic variables.[in] day: The current model time.[in] n: The time step number of the current execution.cs: The control structure returned by a previous call to MOM_sum_output_init.tracer_csp: tracer control structure.obc: Open boundaries control structure.[in] dt_forcing: The forcing time step
-
subroutine, public mom_sum_output::accumulate_net_input(fluxes fluxes, sfc_state sfc_state, dt dt, G G, CS CS) This subroutine accumates the net input of volume, salt and heat, through the ocean surface for use in diagnosing conservation.
- Parameters
[in] fluxes: A structure containing pointers to any possible forcing fields. Unused fields are unallocated.[in] sfc_state: A structure containing fields that describe the surface state of the ocean.[in] dt: The amount of time over which to average [s].[in] g: The ocean’s grid structure.cs: The control structure returned by a previous call to MOM_sum_output_init.
-
subroutine
depth_list_setup(G G, US US, CS CS)¶ This subroutine sets up an ordered list of depths, along with the cross sectional areas at each depth and the volume of fluid deeper than each depth. This might be read from a previously created file or it might be created anew. (For now only new creation occurs.
- Parameters
[in] g: The ocean’s grid structure[in] us: A dimensional unit scaling typecs: The control structure returned by a previous call to MOM_sum_output_init.
-
subroutine
create_depth_list(G G, CS CS)¶ create_depth_list makes an ordered list of depths, along with the cross sectional areas at each depth and the volume of fluid deeper than each depth.
- Parameters
[in] g: The ocean’s grid structure.cs: The control structure set up in MOM_sum_output_init, in which the ordered depth list is stored.
-
subroutine
write_depth_list(G G, US US, CS CS, filename filename, list_size list_size)¶ This subroutine writes out the depth list to the specified file.
- Parameters
[in] g: The ocean’s grid structure.[in] us: A dimensional unit scaling typecs: The control structure returned by a previous call to MOM_sum_output_init.[in] filename: The path to the depth list file to write.[in] list_size: The size of the depth list.
-
subroutine
read_depth_list(G G, US US, CS CS, filename filename)¶ This subroutine reads in the depth list to the specified file and allocates and sets up CSDL and CSlist_size .
- Parameters
[in] g: The ocean’s grid structure[in] us: A dimensional unit scaling typecs: The control structure returned by a previous call to MOM_sum_output_init.[in] filename: The path to the depth list file to read.
-
subroutine
get_depth_list_checksums(G G, depth_chksum depth_chksum, area_chksum area_chksum)¶ Return the checksums required to verify DEPTH_LIST_FILE contents.
This function computes checksums for the bathymetry (GbathyT) and masked area (mask2dT * areaT) fields of the model grid G, which are used to compute the depth list. A difference in checksum indicates that a different method was used to compute the grid data, and that any results using the depth list, such as APE, will not be reproducible.
Checksums are saved as hexadecimal strings, in order to avoid potential datatype issues with netCDF attributes.
- Parameters
[in] g: Ocean grid structure[out] depth_chksum: Depth checksum hexstring[out] area_chksum: Area checksum hexstring
Variables
-
integer, parameter mom_sum_output::num_fields = 17 Number of diagnostic fields.
-
character (*), parameter mom_sum_output::depth_chksum_attr = "bathyT_checksum" Checksum attribute name of GbathyT over the compute domain.
-
character (*), parameter mom_sum_output::area_chksum_attr = "mask2dT_areaT_checksum" Checksum attribute of name of Gmask2dT * GareaT over the compute domain.
-
-
namespace
mom_surface_forcing¶ Functions that calculate the surface wind stresses and fluxes of buoyancy or temperature/salinity andfresh water, in ocean-only (solo) mode.
These functions are called every time step, even if the wind stresses or buoyancy fluxes are constant in time - in that case these routines return quickly without doing anything. In addition, any I/O of forcing fields is controlled by surface_forcing_init, located in this file.
Unnamed Group
-
integer
id_clock_forcing¶ A CPU time clock.
-
subroutine, public mom_surface_forcing::set_forcing(sfc_state sfc_state, forces forces, fluxes fluxes, day_start day_start, day_interval day_interval, G G, US US, CS CS) Calls subroutines in this file to get surface forcing fields.
It also allocates and initializes the fields in the forcing and mech_forcing types the first time it is called.
- Parameters
[inout] sfc_state: A structure containing fields that describe the surface state of the ocean.[inout] forces: A structure with the driving mechanical forces[inout] fluxes: A structure containing thermodynamic forcing fields[in] day_start: The start time of the fluxes[in] day_interval: Length of time over which these fluxes applied[inout] g: The ocean’s grid structure[in] us: A dimensional unit scaling typecs: pointer to control struct returned by a previous surface_forcing_init call
-
subroutine
wind_forcing_const(sfc_state sfc_state, forces forces, tau_x0 tau_x0, tau_y0 tau_y0, day day, G G, US US, CS CS)¶ Sets the surface wind stresses to constant values.
- Parameters
[inout] sfc_state: A structure containing fields that describe the surface state of the ocean.[inout] forces: A structure with the driving mechanical forces[in] tau_x0: The zonal wind stress [Pa][in] tau_y0: The meridional wind stress [Pa][in] day: The time of the fluxes[in] g: The ocean’s grid structure[in] us: A dimensional unit scaling typecs: pointer to control struct returned by a previous surface_forcing_init call
-
subroutine
wind_forcing_2gyre(sfc_state sfc_state, forces forces, day day, G G, US US, CS CS)¶ Sets the surface wind stresses to set up two idealized gyres.
- Parameters
[inout] sfc_state: A structure containing fields that describe the surface state of the ocean.[inout] forces: A structure with the driving mechanical forces[in] day: The time of the fluxes[in] g: The ocean’s grid structure[in] us: A dimensional unit scaling typecs: pointer to control struct returned by a previous surface_forcing_init call
-
subroutine
wind_forcing_1gyre(sfc_state sfc_state, forces forces, day day, G G, US US, CS CS)¶ Sets the surface wind stresses to set up a single idealized gyre.
- Parameters
[inout] sfc_state: A structure containing fields that describe the surface state of the ocean.[inout] forces: A structure with the driving mechanical forces[in] day: The time of the fluxes[in] g: The ocean’s grid structure[in] us: A dimensional unit scaling typecs: pointer to control struct returned by a previous surface_forcing_init call
-
subroutine
wind_forcing_gyres(sfc_state sfc_state, forces forces, day day, G G, US US, CS CS)¶ Sets the surface wind stresses to set up idealized gyres.
- Parameters
[inout] sfc_state: A structure containing fields that describe the surface state of the ocean.[inout] forces: A structure with the driving mechanical forces[in] day: The time of the fluxes[in] g: The ocean’s grid structure[in] us: A dimensional unit scaling typecs: pointer to control struct returned by a previous surface_forcing_init call
-
subroutine
wind_forcing_from_file(sfc_state sfc_state, forces forces, day day, G G, US US, CS CS)¶ - Parameters
[inout] sfc_state: A structure containing fields that describe the surface state of the ocean.[inout] forces: A structure with the driving mechanical forces[in] day: The time of the fluxes[inout] g: The ocean’s grid structure[in] us: A dimensional unit scaling typecs: pointer to control struct returned by a previous surface_forcing_init call
-
subroutine
wind_forcing_by_data_override(sfc_state sfc_state, forces forces, day day, G G, US US, CS CS)¶ - Parameters
[inout] sfc_state: A structure containing fields that describe the surface state of the ocean.[inout] forces: A structure with the driving mechanical forces[in] day: The time of the fluxes[inout] g: The ocean’s grid structure[in] us: A dimensional unit scaling typecs: pointer to control struct returned by a previous surface_forcing_init call
-
subroutine
buoyancy_forcing_from_files(sfc_state sfc_state, fluxes fluxes, day day, dt dt, G G, US US, CS CS)¶ Specifies zero surface bouyancy fluxes from input files.
- Parameters
[inout] sfc_state: A structure containing fields that describe the surface state of the ocean.[inout] fluxes: A structure containing thermodynamic forcing fields[in] day: The time of the fluxes[in] dt: The amount of time over which the fluxes apply [s][inout] g: The ocean’s grid structure[in] us: A dimensional unit scaling typecs: pointer to control struct returned by a previous surface_forcing_init call
-
subroutine
buoyancy_forcing_from_data_override(sfc_state sfc_state, fluxes fluxes, day day, dt dt, G G, US US, CS CS)¶ Specifies zero surface bouyancy fluxes from data over-ride.
- Parameters
[inout] sfc_state: A structure containing fields that describe the surface state of the ocean.[inout] fluxes: A structure containing thermodynamic forcing fields[in] day: The time of the fluxes[in] dt: The amount of time over which the fluxes apply [s][inout] g: The ocean’s grid structure[in] us: A dimensional unit scaling typecs: pointer to control struct returned by a previous surface_forcing_init call
-
subroutine
buoyancy_forcing_zero(sfc_state sfc_state, fluxes fluxes, day day, dt dt, G G, CS CS)¶ This subroutine specifies zero surface bouyancy fluxes.
- Parameters
[inout] sfc_state: A structure containing fields that describe the surface state of the ocean.[inout] fluxes: A structure containing thermodynamic forcing fields[in] day: The time of the fluxes[in] dt: The amount of time over which the fluxes apply [s][in] g: The ocean’s grid structurecs: pointer to control struct returned by a previous surface_forcing_init call
-
subroutine
buoyancy_forcing_const(sfc_state sfc_state, fluxes fluxes, day day, dt dt, G G, CS CS)¶ Sets up spatially and temporally constant surface heat fluxes.
- Parameters
[inout] sfc_state: A structure containing fields that describe the surface state of the ocean.[inout] fluxes: A structure containing thermodynamic forcing fields[in] day: The time of the fluxes[in] dt: The amount of time over which the fluxes apply [s][in] g: The ocean’s grid structurecs: pointer to control struct returned by a previous surface_forcing_init call
-
subroutine
buoyancy_forcing_linear(sfc_state sfc_state, fluxes fluxes, day day, dt dt, G G, CS CS)¶ Sets surface fluxes of heat and salinity by restoring to temperature and salinity profiles that vary linearly with latitude.
- Parameters
[inout] sfc_state: A structure containing fields that describe the surface state of the ocean.[inout] fluxes: A structure containing thermodynamic forcing fields[in] day: The time of the fluxes[in] dt: The amount of time over which the fluxes apply [s][in] g: The ocean’s grid structurecs: pointer to control struct returned by a previous surface_forcing_init call
-
subroutine, public mom_surface_forcing::forcing_save_restart(CS CS, G G, Time Time, directory directory, time_stamped time_stamped, filename_suffix filename_suffix) Save a restart file for the forcing fields.
- Parameters
cs: pointer to control struct returned by a previous surface_forcing_init call[inout] g: The ocean’s grid structure[in] time: model time at this call; needed for mpp_write calls[in] directory: directory into which to write these restart files[in] time_stamped: If true, the restart file names include a unique time stamp; the default is false.[in] filename_suffix: optional suffix (e.g., a time-stamp) to append to the restart fname
-
subroutine, public mom_surface_forcing::surface_forcing_init(Time Time, G G, US US, param_file param_file, diag diag, CS CS, tracer_flow_CSp tracer_flow_CSp) Initialize the surface forcing module.
- Parameters
[in] time: The current model time[in] g: The ocean’s grid structure[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters[inout] diag: structure used to regulate diagnostic outputcs: pointer to control struct returned by a previous surface_forcing_init calltracer_flow_csp: Forcing for tracers?
-
integer
-
namespace
mom_tfreeze¶ Freezing point expressions.
Functions
-
subroutine
calculate_tfreeze_linear_scalar(S S, pres pres, T_Fr T_Fr, TFr_S0_P0 TFr_S0_P0, dTFr_dS dTFr_dS, dTFr_dp dTFr_dp)¶ This subroutine computes the freezing point potential temperature [degC] from salinity [ppt], and pressure [Pa] using a simple linear expression, with coefficients passed in as arguments.
- Parameters
[in] s: salinity [ppt].[in] pres: pressure [Pa].[out] t_fr: Freezing point potential temperature [degC].[in] tfr_s0_p0: The freezing point at S=0, p=0 [degC].[in] dtfr_ds: The derivative of freezing point with salinity, [degC ppt-1].[in] dtfr_dp: The derivative of freezing point with pressure, [degC Pa-1].
-
subroutine
calculate_tfreeze_linear_array(S S, pres pres, T_Fr T_Fr, start start, npts npts, TFr_S0_P0 TFr_S0_P0, dTFr_dS dTFr_dS, dTFr_dp dTFr_dp)¶ This subroutine computes an array of freezing point potential temperatures [degC] from salinity [ppt], and pressure [Pa] using a simple linear expression, with coefficients passed in as arguments.
- Parameters
[in] s: salinity [ppt].[in] pres: pressure [Pa].[out] t_fr: Freezing point potential temperature [degC].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.[in] tfr_s0_p0: The freezing point at S=0, p=0, [degC].[in] dtfr_ds: The derivative of freezing point with salinity, [degC PSU-1].[in] dtfr_dp: The derivative of freezing point with pressure, [degC Pa-1].
-
subroutine
calculate_tfreeze_millero_scalar(S S, pres pres, T_Fr T_Fr)¶ This subroutine computes the freezing point potential temperature [degC] from salinity [ppt], and pressure [Pa] using the expression from Millero (1978) (and in appendix A of Gill 1982), but with the of the pressure dependence changed from 7.53e-8 to 7.75e-8 to make this an expression for potential temperature (not in situ temperature), using a value that is correct at the freezing point at 35 PSU and 5e6 Pa (500 dbar).
- Parameters
[in] s: Salinity in PSU.[in] pres: Pressure [Pa].[out] t_fr: Freezing point potential temperature [degC].
-
subroutine
calculate_tfreeze_millero_array(S S, pres pres, T_Fr T_Fr, start start, npts npts)¶ This subroutine computes the freezing point potential temperature [degC] from salinity [ppt], and pressure [Pa] using the expression from Millero (1978) (and in appendix A of Gill 1982), but with the of the pressure dependence changed from 7.53e-8 to 7.75e-8 to make this an expression for potential temperature (not in situ temperature), using a value that is correct at the freezing point at 35 PSU and 5e6 Pa (500 dbar).
- Parameters
[in] s: Salinity [PSU].[in] pres: Pressure [Pa].[out] t_fr: Freezing point potential temperature [degC].[in] start: The starting point in the arrays.[in] npts: The number of values to calculate.
-
subroutine
calculate_tfreeze_teos10_scalar(S S, pres pres, T_Fr T_Fr)¶ This subroutine computes the freezing point conservative temperature [degC] from absolute salinity [g/kg], and pressure [Pa] using the TEOS10 package.
- Parameters
[in] s: Absolute salinity [g/kg].[in] pres: Pressure [Pa].[out] t_fr: Freezing point conservative temperature [degC].
-
subroutine
calculate_tfreeze_teos10_array(S S, pres pres, T_Fr T_Fr, start start, npts npts)¶ This subroutine computes the freezing point conservative temperature [degC] from absolute salinity [g/kg], and pressure [Pa] using the TEOS10 package.
- Parameters
[in] s: absolute salinity [g/kg].[in] pres: pressure [Pa].[out] t_fr: Freezing point conservative temperature [degC].[in] start: the starting point in the arrays.[in] npts: the number of values to calculate.
-
subroutine
-
namespace
mom_thickness_diffuse¶ Thickness diffusion (or Gent McWilliams)
Functions
-
subroutine, public mom_thickness_diffuse::thickness_diffuse(h h, uhtr uhtr, vhtr vhtr, tv tv, dt dt, G G, GV GV, US US, MEKE MEKE, VarMix VarMix, CDp CDp, CS CS) Calculates thickness diffusion coefficients and applies thickness diffusion to layer thicknesses, h. Diffusivities are limited to ensure stability. Also returns along-layer mass fluxes used in the continuity equation.
- Parameters
[in] g: Ocean grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[inout] h: Layer thickness [H ~> m or kg m-2][inout] uhtr: Accumulated zonal mass flux [m2 H ~> m3 or kg][inout] vhtr: Accumulated meridional mass flux [m2 H ~> m3 or kg][in] tv: Thermodynamics structure[in] dt: Time increment [s]meke: MEKE control structurevarmix: Variable mixing coefficients[inout] cdp: Diagnostics for the continuity equationcs: Control structure for thickness diffusion
-
subroutine
thickness_diffuse_full(h h, e e, Kh_u Kh_u, Kh_v Kh_v, tv tv, uhD uhD, vhD vhD, cg1 cg1, dt dt, G G, GV GV, US US, MEKE MEKE, CS CS, int_slope_u int_slope_u, int_slope_v int_slope_v, slope_x slope_x, slope_y slope_y)¶ Calculates parameterized layer transports for use in the continuity equation. Fluxes are limited to give positive definite thicknesses. Called by thickness_diffuse().
- Parameters
[in] g: Ocean grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[in] h: Layer thickness [H ~> m or kg m-2][in] e: Interface positions [Z ~> m][in] kh_u: Thickness diffusivity on interfaces at u points [m2 s-1][in] kh_v: Thickness diffusivity on interfaces at v points [m2 s-1][in] tv: Thermodynamics structure[out] uhd: Zonal mass fluxes [H m2 s-1 ~> m3 s-1 or kg s-1][out] vhd: Meridional mass fluxes [H m2 s-1 ~> m3 s-1 or kg s-1]cg1: Wave speed [m s-1][in] dt: Time increment [s]meke: MEKE control structurecs: Control structure for thickness diffusion[in] int_slope_u: Ratio that determine how much of the isopycnal slopes are taken directly from the interface slopes without consideration of density gradients.[in] int_slope_v: Ratio that determine how much of the isopycnal slopes are taken directly from the interface slopes without consideration of density gradients.[in] slope_x: Isopycnal slope at u-points[in] slope_y: Isopycnal slope at v-points
-
subroutine
streamfn_solver(nk nk, c2_h c2_h, hN2 hN2, sfn sfn)¶ Tridiagonal solver for streamfunction at interfaces.
- Parameters
[in] nk: Number of layers[in] c2_h: Wave speed squared over thickness in layers [m s-2][in] hn2: Thickness times N2 at interfaces [m s-2][inout] sfn: Streamfunction [Z m2 s-1 ~> m3 s-1] or arbitrary units On entry, equals diffusivity times slope. On exit, equals the streamfunction.
-
subroutine
add_detangling_kh(h h, e e, Kh_u Kh_u, Kh_v Kh_v, KH_u_CFL KH_u_CFL, KH_v_CFL KH_v_CFL, tv tv, dt dt, G G, GV GV, US US, CS CS, int_slope_u int_slope_u, int_slope_v int_slope_v)¶ Modifies thickness diffusivities to untangle layer structures.
- Parameters
[in] g: Ocean grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[in] h: Layer thickness [H ~> m or kg m-2][in] e: Interface positions [Z ~> m][inout] kh_u: Thickness diffusivity on interfaces at u points [m2 s-1][inout] kh_v: Thickness diffusivity on interfaces at v points [m2 s-1][in] kh_u_cfl: Maximum stable thickness diffusivity at u points [m2 s-1][in] kh_v_cfl: Maximum stable thickness diffusivity at v points [m2 s-1][in] tv: Thermodynamics structure[in] dt: Time increment [s]cs: Control structure for thickness diffusion[inout] int_slope_u: Ratio that determine how much of the isopycnal slopes are taken directly from the interface slopes without consideration of density gradients.[inout] int_slope_v: Ratio that determine how much of the isopycnal slopes are taken directly from the interface slopes without consideration of density gradients.
-
subroutine, public mom_thickness_diffuse::thickness_diffuse_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, CDp CDp, CS CS) Initialize the thickness diffusion module/structure.
- Parameters
[in] time: Current model time[in] g: Ocean grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: Parameter file handles[inout] diag: Diagnostics control structure[inout] cdp: Continuity equation diagnosticscs: Control structure for thickness diffusion
-
subroutine, public mom_thickness_diffuse::thickness_diffuse_get_kh(CS CS, KH_u_GME KH_u_GME, KH_v_GME KH_v_GME, G G) Copies ubtav and vbtav from private type into arrays.
- Parameters
cs: Control structure for this module[in] g: Grid structure[inout] kh_u_gme: interface height diffusivities in u-columns [m2 s-1][inout] kh_v_gme: interface height diffusivities in v-columns [m2 s-1]
-
subroutine, public mom_thickness_diffuse::thickness_diffuse_end(CS CS) Deallocate the thickness diffusion control structure.
- Parameters
cs: Control structure for thickness diffusion
-
-
namespace
mom_tidal_forcing¶ Tidal contributions to geopotential.
Functions
-
subroutine, public mom_tidal_forcing::tidal_forcing_init(Time Time, G G, param_file param_file, CS CS) This subroutine allocates space for the static variables used by this module. The metrics may be effectively 0, 1, or 2-D arrays, while fields like the background viscosities are 2-D arrays. ALLOC is a macro defined in MOM_memory.h for allocate or nothing with static memory.
- Parameters
[in] time: The current model time.[inout] g: The ocean’s grid structure.[in] param_file: A structure to parse for run-time parameters.cs: A pointer that is set to point to the control structure for this module.
-
subroutine
find_in_files(filenames filenames, varname varname, array array, G G)¶ This subroutine finds a named variable in a list of files and reads its values into a domain-decomposed 2-d array.
- Parameters
[in] filenames: The names of the files to search for the named variable[in] varname: The name of the variable to read[in] g: The ocean’s grid structure[out] array: The array to fill with the data
-
subroutine, public mom_tidal_forcing::tidal_forcing_sensitivity(G G, CS CS, deta_tidal_deta deta_tidal_deta) This subroutine calculates returns the partial derivative of the local geopotential height with the input sea surface height due to self-attraction and loading.
- Parameters
[in] g: The ocean’s grid structure.cs: The control structure returned by a previous call to tidal_forcing_init.[out] deta_tidal_deta: The partial derivative of eta_tidal with the local value of eta [nondim].
-
subroutine, public mom_tidal_forcing::calc_tidal_forcing(Time Time, eta eta, eta_tidal eta_tidal, G G, CS CS, deta_tidal_deta deta_tidal_deta, m_to_Z m_to_Z) This subroutine calculates the geopotential anomalies that drive the tides, including self-attraction and loading. Optionally, it also returns the partial derivative of the local geopotential height with the input sea surface height. For now, eta and eta_tidal are both geopotential heights in depth units, but probably the input for eta should really be replaced with the column mass anomalies.
- Parameters
[in] g: The ocean’s grid structure.[in] time: The time for the caluculation.[in] eta: The sea surface height anomaly from a time-mean geoid [Z ~> m].[out] eta_tidal: The tidal forcing geopotential height anomalies [Z ~> m].cs: The control structure returned by a previous call to tidal_forcing_init.[out] deta_tidal_deta: The partial derivative of eta_tidal with the local value of eta [nondim].[in] m_to_z: A scaling factor from m to the units of eta.
-
subroutine, public mom_tidal_forcing::tidal_forcing_end(CS CS) This subroutine deallocates memory associated with the tidal forcing module.
- Parameters
cs: The control structure returned by a previous call to tidal_forcing_init; it is deallocated here.
Variables
-
integer, parameter mom_tidal_forcing::max_constituents = 10 The maximum number of tidal constituents that could be used.
-
integer
id_clock_tides¶ CPU clock for tides.
-
-
namespace
mom_tidal_mixing¶ Interface to vertical tidal mixing schemes including CVMix tidal mixing.
Unnamed Group
-
character*(20), parameter mom_tidal_mixing::stlaurent_profile_string = "STLAURENT_02" Initializes internal tidal dissipation scheme for diapycnal mixing.
- Parameters
[in] time: The current time.[in] g: Grid structure.[in] gv: Vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: Run-time parameter file handle[inout] diag: Diagnostics control structure.cs: This module’s control structure.
-
character*(20), parameter mom_tidal_mixing::polzin_profile_string = "POLZIN_09" Initializes internal tidal dissipation scheme for diapycnal mixing.
- Parameters
[in] time: The current time.[in] g: Grid structure.[in] gv: Vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: Run-time parameter file handle[inout] diag: Diagnostics control structure.cs: This module’s control structure.
-
integer, parameter mom_tidal_mixing::stlaurent_02 = 1 Initializes internal tidal dissipation scheme for diapycnal mixing.
- Parameters
[in] time: The current time.[in] g: Grid structure.[in] gv: Vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: Run-time parameter file handle[inout] diag: Diagnostics control structure.cs: This module’s control structure.
-
integer, parameter mom_tidal_mixing::polzin_09 = 2 Initializes internal tidal dissipation scheme for diapycnal mixing.
- Parameters
[in] time: The current time.[in] g: Grid structure.[in] gv: Vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: Run-time parameter file handle[inout] diag: Diagnostics control structure.cs: This module’s control structure.
-
character*(20), parameter mom_tidal_mixing::simmons_scheme_string = "SIMMONS" Initializes internal tidal dissipation scheme for diapycnal mixing.
- Parameters
[in] time: The current time.[in] g: Grid structure.[in] gv: Vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: Run-time parameter file handle[inout] diag: Diagnostics control structure.cs: This module’s control structure.
-
character*(20), parameter mom_tidal_mixing::schmittner_scheme_string = "SCHMITTNER" Initializes internal tidal dissipation scheme for diapycnal mixing.
- Parameters
[in] time: The current time.[in] g: Grid structure.[in] gv: Vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: Run-time parameter file handle[inout] diag: Diagnostics control structure.cs: This module’s control structure.
-
integer, parameter mom_tidal_mixing::simmons = 1 Initializes internal tidal dissipation scheme for diapycnal mixing.
- Parameters
[in] time: The current time.[in] g: Grid structure.[in] gv: Vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: Run-time parameter file handle[inout] diag: Diagnostics control structure.cs: This module’s control structure.
-
integer, parameter mom_tidal_mixing::schmittner = 2 Initializes internal tidal dissipation scheme for diapycnal mixing.
- Parameters
[in] time: The current time.[in] g: Grid structure.[in] gv: Vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: Run-time parameter file handle[inout] diag: Diagnostics control structure.cs: This module’s control structure.
-
logical function, public mom_tidal_mixing::tidal_mixing_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS) Initializes internal tidal dissipation scheme for diapycnal mixing.
- Parameters
[in] time: The current time.[in] g: Grid structure.[in] gv: Vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: Run-time parameter file handle[inout] diag: Diagnostics control structure.cs: This module’s control structure.
-
subroutine, public mom_tidal_mixing::calculate_tidal_mixing(h h, N2_bot N2_bot, j j, TKE_to_Kd TKE_to_Kd, max_TKE max_TKE, G G, GV GV, US US, CS CS, N2_lay N2_lay, N2_int N2_int, Kd_lay Kd_lay, Kd_int Kd_int, Kd_max Kd_max, Kv Kv) Depending on whether or not CVMix is active, calls the associated subroutine to compute internal tidal dissipation and to add the effect of internal-tide-driven mixing to the layer or interface diffusivities.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[in] h: Layer thicknesses [H ~> m or kg m-2][in] n2_bot: The near-bottom squared buoyancy frequency [T-2 ~> s-2].[in] n2_lay: The squared buoyancy frequency of the layers [T-2 ~> s-2].[in] n2_int: The squared buoyancy frequency at the interfaces [T-2 ~> s-2].[in] j: The j-index to work on[in] tke_to_kd: The conversion rate between the TKE dissipated within a layer and the diapycnal diffusivity within that layer, usually (~Rho_0 / (G_Earth * dRho_lay)) [Z2 T-1 / Z3 T-3 = T2 Z-1 ~> s2 m-1][in] max_tke: The energy required to for a layer to entrain to its maximum realizable thickness [Z3 T-3 ~> m3 s-3]cs: The control structure for this module[inout] kd_lay: The diapycnal diffusvity in layers [Z2 T-1 ~> m2 s-1].[inout] kd_int: The diapycnal diffusvity at interfaces,[in] kd_max: The maximum increment for diapycnal diffusivity due to TKE-based processes, [Z2 T-1 ~> m2 s-1]. Set this to a negative value to have no limit.kv: The “slow” vertical viscosity at each interface (not layer!) [Z2 T-1 ~> m2 s-1].
-
subroutine
calculate_cvmix_tidal(h h, j j, G G, GV GV, US US, CS CS, N2_int N2_int, Kd_lay Kd_lay, Kv Kv)¶ Calls the CVMix routines to compute tidal dissipation and to add the effect of internal-tide-driven mixing to the interface diffusivities.
- Parameters
[in] j: The j-index to work on[in] g: Grid structure.[in] gv: ocean vertical grid structure[in] us: A dimensional unit scaling typecs: This module’s control structure.[in] n2_int: The squared buoyancy frequency at the interfaces [T-2 ~> s-2].[in] h: Layer thicknesses [H ~> m or kg m-2].[inout] kd_lay: The diapycnal diffusivities in the layers [Z2 T-1 ~> m2 s-1].kv: The “slow” vertical viscosity at each interface (not layer!) [Z2 T-1 ~> m2 s-1].
-
subroutine
add_int_tide_diffusivity(h h, N2_bot N2_bot, j j, TKE_to_Kd TKE_to_Kd, max_TKE max_TKE, G G, GV GV, US US, CS CS, N2_lay N2_lay, Kd_lay Kd_lay, Kd_int Kd_int, Kd_max Kd_max)¶ This subroutine adds the effect of internal-tide-driven mixing to the layer diffusivities. The mechanisms considered are (1) local dissipation of internal waves generated by the barotropic flow (“itidal”), (2) local dissipation of internal waves generated by the propagating low modes (rays) of the internal tide (“lowmode”), and (3) local dissipation of internal lee waves. Will eventually need to add diffusivity due to other wave-breaking processes (e.g. Bottom friction, Froude-number-depending breaking, PSI, etc.).
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[in] h: Layer thicknesses [H ~> m or kg m-2][in] n2_bot: The near-bottom squared buoyancy frequency frequency [T-2 ~> s-2].[in] n2_lay: The squared buoyancy frequency of the layers [T-2 ~> s-2].[in] j: The j-index to work on[in] tke_to_kd: The conversion rate between the TKE dissipated within a layer and the diapycnal diffusivity within that layer, usually (~Rho_0 / (G_Earth * dRho_lay)) [Z2 T-1 / Z3 T-3 = T2 Z-1 ~> s2 m-1][in] max_tke: The energy required to for a layer to entrain to its maximum realizable thickness [Z3 T-3 ~> m3 s-3]cs: The control structure for this module[inout] kd_lay: The diapycnal diffusvity in layers [Z2 T-1 ~> m2 s-1].[inout] kd_int: The diapycnal diffusvity at interfaces[in] kd_max: The maximum increment for diapycnal diffusivity due to TKE-based processes [Z2 T-1 ~> m2 s-1]. Set this to a negative value to have no limit.
-
subroutine, public mom_tidal_mixing::setup_tidal_diagnostics(G G, CS CS) Sets up diagnostics arrays for tidal mixing.
- Parameters
[in] g: The ocean’s grid structurecs: The control structure for this module
-
subroutine, public mom_tidal_mixing::post_tidal_diagnostics(G G, GV GV, h h, CS CS) This subroutine offers up diagnostics of the tidal mixing.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure.[in] h: Layer thicknesses [H ~> m or kg m-2].cs: The control structure for this module
-
subroutine
read_tidal_energy(G G, US US, tidal_energy_type tidal_energy_type, tidal_energy_file tidal_energy_file, CS CS)¶ This subroutine read tidal energy inputs from a file.
- Parameters
[in] g: The ocean’s grid structure[in] us: A dimensional unit scaling type[in] tidal_energy_type: The type of tidal energy inputs to read[in] tidal_energy_file: The file from which to read tidalinputscs: The control structure for this module
-
subroutine
read_tidal_constituents(G G, US US, tidal_energy_file tidal_energy_file, CS CS)¶ This subroutine reads tidal input energy from a file by constituent.
- Parameters
[in] g: The ocean’s grid structure[in] us: A dimensional unit scaling type[in] tidal_energy_file: The file from which to read tidal energy inputscs: The control structure for this module
-
subroutine, public mom_tidal_mixing::tidal_mixing_end(CS CS) Clear pointers and deallocate memory.
- Parameters
cs: This module’s control structure, which will be deallocated in this routine.
-
-
namespace
mom_time_manager¶ Wraps the FMS time manager functions.
Functions
-
type(time_type) function, public mom_time_manager::real_to_time(x x, err_msg err_msg) This is an alternate implementation of the FMS function real_to_time_type that is accurate over a larger range of input values. With 32 bit signed integers, this version should work over the entire valid range (2^31 days or ~5.8835 million years) of time_types, whereas the standard version in the FMS time_manager stops working for conversions of times greater than 2^31 seconds, or ~68.1 years.
- Return
The output time as a time_type
- Parameters
[in] x: The input time in real seconds.[out] err_msg: An optional returned error message.
-
-
namespace
mom_tracer_advect¶ This program contains the subroutines that advect tracers along coordinate surfaces.
This program contains the subroutines that advect tracers horizontally (i.e. along layers).
Unnamed Group
-
integer
id_clock_advect¶ CPU time clocks.
-
integer
id_clock_pass¶ CPU time clocks.
-
integer
id_clock_sync¶ CPU time clocks.
-
subroutine, public mom_tracer_advect::advect_tracer(h_end h_end, uhtr uhtr, vhtr vhtr, OBC OBC, dt dt, G G, GV GV, CS CS, Reg Reg, h_prev_opt h_prev_opt, max_iter_in max_iter_in, x_first_in x_first_in, uhr_out uhr_out, vhr_out vhr_out, h_out h_out) This routine time steps the tracer concentration using a monotonic, conservative, weakly diffusive scheme.
- Parameters
[inout] g: ocean grid structure[in] gv: ocean vertical grid structure[in] h_end: layer thickness after advection [H ~> m or kg m-2][in] uhtr: accumulated volume/mass flux through zonal face [H m2 ~> m3 or kg][in] vhtr: accumulated volume/mass flux through merid face [H m2 ~> m3 or kg]obc: specifies whether, where, and what OBCs are used[in] dt: time increment [s]cs: control structure for modulereg: pointer to tracer registry[in] h_prev_opt: layer thickness before advection [H ~> m or kg m-2][in] max_iter_in: The maximum number of iterations[in] x_first_in: If present, indicate whether to update first in the x- or y-direction.[out] uhr_out: accumulated volume/mass flux through zonal face[out] vhr_out: accumulated volume/mass flux through merid face[out] h_out: layer thickness before advection [H ~> m or kg m-2]
-
subroutine
advect_x(Tr Tr, hprev hprev, uhr uhr, uh_neglect uh_neglect, OBC OBC, domore_u domore_u, ntr ntr, Idt Idt, is is, ie ie, js js, je je, k k, G G, GV GV, usePPM usePPM, useHuynh useHuynh)¶ This subroutine does 1-d flux-form advection in the zonal direction using a monotonic piecewise linear scheme.
- Parameters
[inout] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[inout] tr: The array of registered tracers to work on[inout] hprev: cell volume at the end of previous tracer change [H m2 ~> m3 or kg][inout] uhr: accumulated volume/mass flux through the zonal face [H m2 ~> m3 or kg][inout] uh_neglect: A tiny zonal mass flux that can be neglected [H m2 ~> m3 or kg]obc: specifies whether, where, and what OBCs are used[inout] domore_u: If true, there is more advection to be done in this u-row[in] idt: The inverse of dt [s-1][in] ntr: The number of tracers[in] is: The starting tracer i-index to work on[in] ie: The ending tracer i-index to work on[in] js: The starting tracer j-index to work on[in] je: The ending tracer j-index to work on[in] k: The k-level to work on[in] useppm: If true, use PPM instead of PLM[in] usehuynh: If true, use the Huynh scheme for PPM interface values
-
subroutine
advect_y(Tr Tr, hprev hprev, vhr vhr, vh_neglect vh_neglect, OBC OBC, domore_v domore_v, ntr ntr, Idt Idt, is is, ie ie, js js, je je, k k, G G, GV GV, usePPM usePPM, useHuynh useHuynh)¶ This subroutine does 1-d flux-form advection using a monotonic piecewise linear scheme.
- Parameters
[inout] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[inout] tr: The array of registered tracers to work on[inout] hprev: cell volume at the end of previous tracer change [H m2 ~> m3 or kg][inout] vhr: accumulated volume/mass flux through the meridional face [H m2 ~> m3 or kg][inout] vh_neglect: A tiny meridional mass flux that can be neglected [H m2 ~> m3 or kg]obc: specifies whether, where, and what OBCs are used[inout] domore_v: If true, there is more advection to be done in this v-row[in] idt: The inverse of dt [s-1][in] ntr: The number of tracers[in] is: The starting tracer i-index to work on[in] ie: The ending tracer i-index to work on[in] js: The starting tracer j-index to work on[in] je: The ending tracer j-index to work on[in] k: The k-level to work on[in] useppm: If true, use PPM instead of PLM[in] usehuynh: If true, use the Huynh scheme for PPM interface values
-
subroutine, public mom_tracer_advect::tracer_advect_init(Time Time, G G, param_file param_file, diag diag, CS CS) Initialize lateral tracer advection module.
- Parameters
[in] time: current model time[in] g: ocean grid structure[in] param_file: open file to parse for model parameters[inout] diag: regulates diagnostic outputcs: module control structure
-
subroutine, public mom_tracer_advect::tracer_advect_end(CS CS) Close the tracer advection module.
- Parameters
cs: module control structure
-
integer
-
namespace
mom_tracer_diabatic¶ This module contains routines that implement physical fluxes of tracers (e.g. due to surface fluxes or mixing). These are intended to be called from call_tracer_column_fns in the MOM_tracer_flow_control module.
Functions
-
subroutine, public mom_tracer_diabatic::tracer_vertdiff(h_old h_old, ea ea, eb eb, dt dt, tr tr, G G, GV GV, sfc_flux sfc_flux, btm_flux btm_flux, btm_reservoir btm_reservoir, sink_rate sink_rate, convert_flux_in convert_flux_in) This subroutine solves a tridiagonal equation for the final tracer concentrations after the dual-entrainments, and possibly sinking or surface and bottom sources, are applied. The sinking is implemented with an fully implicit upwind advection scheme.
- Parameters
[in] g: ocean grid structure[in] gv: ocean vertical grid structure[in] h_old: layer thickness before entrainment [H ~> m or kg m-2][in] ea: amount of fluid entrained from the layer above [H ~> m or kg m-2][in] eb: amount of fluid entrained from the layer below [H ~> m or kg m-2][inout] tr: tracer concentration in concentration units [CU][in] dt: amount of time covered by this call [s][in] sfc_flux: surface flux of the tracer [CU kg m-2 s-1][in] btm_flux: The (negative upward) bottom flux of the tracer [CU kg m-2 s-1][inout] btm_reservoir: amount of tracer in a bottom reservoir [CU kg m-2]; formerly [CU m][in] sink_rate: rate at which the tracer sinks [m s-1][in] convert_flux_in: True if the specified sfc_flux needs to be integrated in time
-
subroutine, public mom_tracer_diabatic::applytracerboundaryfluxesinout(G G, GV GV, Tr Tr, dt dt, fluxes fluxes, h h, evap_CFL_limit evap_CFL_limit, minimum_forcing_depth minimum_forcing_depth, in_flux_optional in_flux_optional, out_flux_optional out_flux_optional, update_h_opt update_h_opt) This routine is modeled after applyBoundaryFluxesInOut in MOM_diabatic_aux.F90 NOTE: Please note that in this routine sfc_flux gets set to zero to ensure that the surface flux of the tracer does not get applied again during a subsequent call to tracer_vertdif.
- Parameters
[in] g: Grid structure[in] gv: ocean vertical grid structure[inout] tr: Tracer concentration on T-cell[in] dt: Time-step over which forcing is applied [s][in] fluxes: Surface fluxes container[inout] h: Layer thickness [H ~> m or kg m-2][in] evap_cfl_limit: Limit on the fraction of the water that can be fluxed out of the top layer in a timestep [nondim][in] minimum_forcing_depth: The smallest depth over which fluxes can be applied [m][in] in_flux_optional: The total time-integrated amount of tracer that enters with freshwater[in] out_flux_optional: The total time-integrated amount of tracer that leaves with freshwater[in] update_h_opt: Optional flag to determine whether h should be updated
-
-
namespace
MOM_tracer_flow_control¶ By Will Cooke, April 2003 Edited by Elizabeth Yankovsky, May 2019
This module contains two subroutines into which calls to other tracer initialization (call_tracer_init_fns) and column physics routines (call_tracer_column_fns) can be inserted.
-
namespace
mom_tracer_flow_control¶ Orchestrates the registration and calling of tracer packages.
Unnamed Group
-
subroutine, public mom_tracer_flow_control::call_tracer_flux_init(verbosity verbosity) This subroutine carries out a series of calls to initialize the air-sea tracer fluxes, but it does not record the generated indicies, and it may be called before the ocean model has been initialized and may be called on non-ocean PEs. It is not necessary to call this routine for ocean-only runs, because the same calls are made again inside of the routines called by call_tracer_register.
- Parameters
[in] verbosity: A 0-9 integer indicating a level of verbosity.
-
subroutine, public mom_tracer_flow_control::call_tracer_register(HI HI, GV GV, US US, param_file param_file, CS CS, tr_Reg tr_Reg, restart_CS restart_CS) The following 5 subroutines and associated definitions provide the machinery to register and call the subroutines that initialize tracers and apply vertical column processes to tracers.
- Parameters
[in] hi: A horizontal index type structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters.cs: A pointer that is set to point to the control structure for this module.tr_reg: A pointer that is set to point to the control structure for the tracer advection and diffusion module.restart_cs: A pointer to the restart control structure.
-
subroutine, public mom_tracer_flow_control::tracer_flow_control_init(restart restart, day day, G G, GV GV, US US, h h, param_file param_file, diag diag, OBC OBC, CS CS, sponge_CSp sponge_CSp, ALE_sponge_CSp ALE_sponge_CSp, tv tv) This subroutine calls all registered tracer initialization subroutines.
- Parameters
[in] restart: 1 if the fields have already been read from a restart file.[in] day: Time of the start of the run.[inout] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] h: Layer thicknesses [H ~> m or kg m-2][in] param_file: A structure to parse for run-time parameters[in] diag: A structure that is used to regulate diagnostic output.obc: This open boundary condition type specifies whether, where, and what open boundary conditions are used.cs: The control structure returned by a previous call to call_tracer_register.sponge_csp: A pointer to the control structure for the sponges, if they are in use. Otherwise this may be unassociated.ale_sponge_csp: A pointer to the control structure for the ALE sponges, if they are in use. Otherwise this may be unassociated.[in] tv: A structure pointing to various thermodynamic variables
-
subroutine, public mom_tracer_flow_control::get_chl_from_model(Chl_array Chl_array, G G, CS CS) This subroutine extracts the chlorophyll concentrations from the model state, if possible.
- Parameters
[out] chl_array: The array in which to store the model’s[in] g: The ocean’s grid structure.cs: The control structure returned by a previous call to call_tracer_register.
-
subroutine, public mom_tracer_flow_control::call_tracer_set_forcing(state state, fluxes fluxes, day_start day_start, day_interval day_interval, G G, CS CS) This subroutine calls the individual tracer modules’ subroutines to specify or read quantities related to their surface forcing.
- Parameters
[inout] state: A structure containing fields that describe the surface state of the ocean.[inout] fluxes: A structure containing pointers to any possible forcing fields. Unused fields have NULL ptrs.[in] day_start: Start time of the fluxes.[in] day_interval: Length of time over which these fluxes will be applied.[in] g: The ocean’s grid structure.cs: The control structure returned by a previous call to call_tracer_register.
-
subroutine, public mom_tracer_flow_control::call_tracer_column_fns(h_old h_old, h_new h_new, ea ea, eb eb, fluxes fluxes, Hml Hml, dt dt, G G, GV GV, tv tv, optics optics, CS CS, debug debug, evap_CFL_limit evap_CFL_limit, minimum_forcing_depth minimum_forcing_depth) This subroutine calls all registered tracer column physics subroutines.
- Parameters
[in] h_old: Layer thickness before entrainment [H ~> m or kg m-2].[in] h_new: Layer thickness after entrainment [H ~> m or kg m-2].[in] ea: an array to which the amount of fluid entrained from the layer above during this call will be added [H ~> m or kg m-2].[in] eb: an array to which the amount of fluid entrained from the layer below during this call will be added [H ~> m or kg m-2].[in] fluxes: A structure containing pointers to any possible forcing fields. Unused fields have NULL ptrs.[in] hml: Mixed layer depth [H ~> m or kg m-2][in] dt: The amount of time covered by this call [s][in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] tv: A structure pointing to various thermodynamic variables.optics: The structure containing optical properties.cs: The control structure returned by a previous call to call_tracer_register.[in] debug: If true calculate checksums[in] evap_cfl_limit: Limit on the fraction of the water that can be fluxed out of the top layer in a timestep [nondim][in] minimum_forcing_depth: The smallest depth over which fluxes can be applied [H ~> m or kg m-2]
-
subroutine, public mom_tracer_flow_control::call_tracer_stocks(h h, stock_values stock_values, G G, GV GV, CS CS, stock_names stock_names, stock_units stock_units, num_stocks num_stocks, stock_index stock_index, got_min_max got_min_max, global_min global_min, global_max global_max, xgmin xgmin, ygmin ygmin, zgmin zgmin, xgmax xgmax, ygmax ygmax, zgmax zgmax) This subroutine calls all registered tracer packages to enable them to add to the surface state returned to the coupler. These routines are optional.
- Parameters
[in] h: Layer thicknesses [H ~> m or kg m-2][out] stock_values: The integrated amounts of a tracer on the current PE, usually in kg x concentration [kg conc].[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.cs: The control structure returned by a previous call to call_tracer_register.[out] stock_names: Diagnostic names to use for each stock.[out] stock_units: Units to use in the metadata for each stock.[out] num_stocks: The number of tracer stocks being returned.[in] stock_index: The integer stock index from stocks_constants_mod of the stock to be returned. If this is present and greater than 0, only a single stock can be returned.[inout] got_min_max: Indicates whether the global min and[out] global_min: The global minimum of each tracer[out] global_max: The global maximum of each tracer[out] xgmin: The x-position of the global minimum[out] ygmin: The y-position of the global minimum[out] zgmin: The z-position of the global minimum[out] xgmax: The x-position of the global maximum[out] ygmax: The y-position of the global maximum[out] zgmax: The z-position of the global maximum
-
subroutine
store_stocks(pkg_name pkg_name, ns ns, names names, units units, values values, index index, stock_values stock_values, set_pkg_name set_pkg_name, max_ns max_ns, ns_tot ns_tot, stock_names stock_names, stock_units stock_units)¶ This routine stores the stocks and does error handling for call_tracer_stocks.
- Parameters
[in] pkg_name: The tracer package name[in] ns: The number of stocks associated with this tracer package[in] names: Diagnostic names to use for each stock.[in] units: Units to use in the metadata for each stock.[in] values: The values of the tracer stocks[in] index: The integer stock index from stocks_constants_mod of the stock to be returned. If this is present and greater than 0, only a single stock can be returned.[inout] stock_values: The master list of stock values[inout] set_pkg_name: The name of the last tracer package whose stocks were stored for a specific index. This is used to trigger an error if there are redundant stocks.[in] max_ns: The maximum size of the master stock list[inout] ns_tot: The total number of stocks in the master list[inout] stock_names: Diagnostic names to use for each stock in the master list[inout] stock_units: Units to use in the metadata for each stock in the master list
-
subroutine, public mom_tracer_flow_control::call_tracer_surface_state(state state, h h, G G, CS CS) This subroutine calls all registered tracer packages to enable them to add to the surface state returned to the coupler. These routines are optional.
- Parameters
[inout] state: A structure containing fields that describe the surface state of the ocean.[in] h: Layer thicknesses [H ~> m or kg m-2][in] g: The ocean’s grid structure.cs: The control structure returned by a previous call to call_tracer_register.
-
subroutine, public mom_tracer_flow_control::tracer_flow_control_end(CS CS) - Parameters
cs: The control structure returned by a previous call to call_tracer_register.
-
-
namespace
mom_tracer_hor_diff¶ Main routine for lateral (along surface or neutral) diffusion of tracers.
Unnamed Group
-
integer
id_clock_diffuse¶ CPU time clocks.
-
integer
id_clock_epimix¶ CPU time clocks.
-
integer
id_clock_pass¶ CPU time clocks.
-
integer
id_clock_sync¶ CPU time clocks.
-
subroutine, public mom_tracer_hor_diff::tracer_hordiff(h h, dt dt, MEKE MEKE, VarMix VarMix, G G, GV GV, CS CS, Reg Reg, tv tv, do_online_flag do_online_flag, read_khdt_x read_khdt_x, read_khdt_y read_khdt_y) Compute along-coordinate diffusion of all tracers using the diffusivity in CSKhTr, or using space-dependent diffusivity. Multiple iterations are used (if necessary) so that there is no limit on the acceptable time increment.
- Parameters
[inout] g: Grid type[in] h: Layer thickness [H ~> m or kg m-2][in] dt: time step [s]meke: MEKE typevarmix: Variable mixing type[in] gv: ocean vertical grid structurecs: module control structurereg: registered tracers[in] tv: A structure containing pointers to any available thermodynamic fields, including potential temp and salinity or mixed layer density. Absent fields have NULL ptrs, and these may (probably will) point to some of the same arrays as Tr does. tv is required for epipycnal mixing between mixed layer and the interior.[in] do_online_flag: If present and true, do online tracer transport with stored velcities.[in] read_khdt_x: If present, these are the zonal[in] read_khdt_y: If present, these are the meridional
-
subroutine
tracer_epipycnal_ml_diff(h h, dt dt, Tr Tr, ntr ntr, khdt_epi_x khdt_epi_x, khdt_epi_y khdt_epi_y, G G, GV GV, CS CS, tv tv, num_itts num_itts)¶ This subroutine does epipycnal diffusion of all tracers between the mixed and buffer layers and the interior, using the diffusivity in CSKhTr. Multiple iterations are used (if necessary) so that there is no limit on the acceptable time increment.
- Parameters
[inout] g: ocean grid structure[in] gv: ocean vertical grid structure[in] h: layer thickness [H ~> m or kg m-2][in] dt: time step[inout] tr: tracer array[in] ntr: number of tracers[in] khdt_epi_x: needs a comment[in] khdt_epi_y: needs a comment[inout] cs: module control structure[in] tv: thermodynamic structure[in] num_itts: number of iterations (usually=1)
-
subroutine, public mom_tracer_hor_diff::tracer_hor_diff_init(Time Time, G G, param_file param_file, diag diag, EOS EOS, CS CS) Initialize lateral tracer diffusion module.
- Parameters
[in] time: current model time[in] g: ocean grid structure[inout] diag: diagnostic control[in] eos: Equation of state CS[in] param_file: parameter filecs: horz diffusion control structure
-
subroutine, public mom_tracer_hor_diff::tracer_hor_diff_end(CS CS) - Parameters
cs: module control structure
-
integer
-
namespace
mom_tracer_initialization_from_z¶ Initializes hydrography from z-coordinate climatology files.
Functions
-
subroutine, public mom_tracer_initialization_from_z::mom_initialize_tracer_from_z(h h, tr tr, G G, GV GV, US US, PF PF, src_file src_file, src_var_nam src_var_nam, src_var_unit_conversion src_var_unit_conversion, src_var_record src_var_record, homogenize homogenize, useALEremapping useALEremapping, remappingScheme remappingScheme, src_var_gridspec src_var_gridspec) Initializes a tracer from a z-space data file.
- Parameters
[inout] g: Ocean grid structure.[in] gv: Ocean vertical grid structure.[in] us: A dimensional unit scaling type[in] h: Layer thickness [H ~> m or kg m-2].tr: Pointer to array to be initialized[in] pf: parameter file[in] src_file: source filename[in] src_var_nam: variable name in file[in] src_var_unit_conversion: optional multiplicative unit conversion[in] src_var_record: record to read for multiple time-level files[in] homogenize: optionally homogenize to mean value[in] usealeremapping: to remap or not (optional)[in] remappingscheme: remapping scheme to use.[in] src_var_gridspec: Source variable name in a gridspec file. This is not implemented yet.
Variables
-
character(len=40) mom_tracer_initialization_from_z::mdl = "MOM_tracer_initialization_from_Z" This module’s name.
-
-
namespace
mom_tracer_registry¶ This module contains the tracer_registry_type and the subroutines that handle registration of tracers and related subroutines. The primary subroutine, register_tracer, is called to indicate the tracers advected and diffused.
Unnamed Group
-
subroutine, public mom_tracer_registry::register_tracer(tr_ptr tr_ptr, Reg Reg, param_file param_file, HI HI, GV GV, name name, longname longname, units units, cmor_name cmor_name, cmor_units cmor_units, cmor_longname cmor_longname, tr_desc tr_desc, OBC_inflow OBC_inflow, OBC_in_u OBC_in_u, OBC_in_v OBC_in_v, ad_x ad_x, ad_y ad_y, df_x df_x, df_y df_y, ad_2d_x ad_2d_x, ad_2d_y ad_2d_y, df_2d_x df_2d_x, df_2d_y df_2d_y, advection_xy advection_xy, registry_diags registry_diags, flux_nameroot flux_nameroot, flux_longname flux_longname, flux_units flux_units, flux_scale flux_scale, convergence_units convergence_units, convergence_scale convergence_scale, cmor_tendprefix cmor_tendprefix, diag_form diag_form, restart_CS restart_CS, mandatory mandatory) This subroutine registers a tracer to be advected and laterally diffused.
- Parameters
[in] hi: horizontal index type[in] gv: ocean vertical grid structurereg: pointer to the tracer registrytr_ptr: target or pointer to the tracer array[in] param_file: file to parse for model parameter values[in] name: Short tracer name[in] longname: The long tracer name[in] units: The units of this tracer[in] cmor_name: CMOR name[in] cmor_units: CMOR physical dimensions of variable[in] cmor_longname: CMOR long name[in] tr_desc: A structure with metadata about the tracer[in] obc_inflow: the tracer for all inflows via OBC for which OBC_in_u or OBC_in_v are not specified (units of tracer CONC)obc_in_u: tracer at inflows through u-faces of tracer cells (units of tracer CONC)obc_in_v: tracer at inflows through v-faces of tracer cells (units of tracer CONC)ad_x: diagnostic x-advective flux (CONC m3/s or CONC*kg/s)ad_y: diagnostic y-advective flux (CONC m3/s or CONC*kg/s)df_x: diagnostic x-diffusive flux (CONC m3/s or CONC*kg/s)df_y: diagnostic y-diffusive flux (CONC m3/s or CONC*kg/s)ad_2d_x: vert sum of diagnostic x-advect flux (CONC m3/s or CONC*kg/s)ad_2d_y: vert sum of diagnostic y-advect flux (CONC m3/s or CONC*kg/s)df_2d_x: vert sum of diagnostic x-diffuse flux (CONC m3/s or CONC*kg/s)df_2d_y: vert sum of diagnostic y-diffuse flux (CONC m3/s or CONC*kg/s)advection_xy: convergence of lateral advective tracer fluxes[in] registry_diags: If present and true, use the registry for the diagnostics of this tracer.[in] flux_nameroot: Short tracer name snippet used construct the names of flux diagnostics.[in] flux_longname: A word or phrase used construct the long names of flux diagnostics.[in] flux_units: The units for the fluxes of this tracer.[in] flux_scale: A scaling factor used to convert the fluxes of this tracer to its desired units.[in] convergence_units: The units for the flux convergence of this tracer.[in] convergence_scale: A scaling factor used to convert the flux convergence of this tracer to its desired units.[in] cmor_tendprefix: The CMOR name for the layer-integrated tendencies of this tracer.[in] diag_form: An integer (1 or 2, 1 by default) indicating the character string template to use in labeling diagnosticsrestart_cs: A pointer to the restart control structure this tracer will be registered for restarts if this argument is present[in] mandatory: If true, this tracer must be read from a restart file.
-
subroutine, public mom_tracer_registry::lock_tracer_registry(Reg Reg) This subroutine locks the tracer registry to prevent the addition of more tracers. After locked=.true., can then register common diagnostics.
- Parameters
reg: pointer to the tracer registry
-
subroutine, public mom_tracer_registry::register_tracer_diagnostics(Reg Reg, h h, Time Time, diag diag, G G, GV GV, use_ALE use_ALE) register_tracer_diagnostics does a set of register_diag_field calls for any previously registered in a tracer registry with a value of registry_diags set to .true.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structurereg: pointer to the tracer registry[in] h: Layer thicknesses[in] time: current model time[in] diag: structure to regulate diagnostic output[in] use_ale: If true active diagnostics that only apply to ALE configurations
-
subroutine, public mom_tracer_registry::preale_tracer_diagnostics(Reg Reg, G G, GV GV) - Parameters
reg: pointer to the tracer registry[in] g: The ocean’s grid structure[in] gv: ocean vertical grid structure
-
subroutine, public mom_tracer_registry::postale_tracer_diagnostics(Reg Reg, G G, GV GV, diag diag, dt dt) - Parameters
reg: pointer to the tracer registry[in] g: The ocean’s grid structure[in] gv: ocean vertical grid structure[in] diag: regulates diagnostic output[in] dt: total time interval for these diagnostics
-
subroutine, public mom_tracer_registry::post_tracer_diagnostics(Reg Reg, h h, diag_prev diag_prev, diag diag, G G, GV GV, dt dt) post_tracer_diagnostics does post_data calls for any diagnostics that are being handled via the tracer registry.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structurereg: pointer to the tracer registry[in] h: Layer thicknesses[in] diag_prev: Contains diagnostic grids from previous timestep[inout] diag: structure to regulate diagnostic output[in] dt: total time step for tracer updates
-
subroutine, public mom_tracer_registry::post_tracer_transport_diagnostics(G G, GV GV, Reg Reg, h_diag h_diag, diag diag) Post the advective and diffusive tendencies.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structurereg: pointer to the tracer registry[in] h_diag: Layer thicknesses on which to post fields[in] diag: structure to regulate diagnostic output
-
subroutine, public mom_tracer_registry::mom_tracer_chksum(mesg mesg, Tr Tr, ntr ntr, G G) This subroutine writes out chksums for tracers.
- Parameters
[in] mesg: message that appears on the chksum lines[in] tr: array of all of registered tracers[in] ntr: number of registered tracers[in] g: ocean grid structure
-
subroutine, public mom_tracer_registry::mom_tracer_chkinv(mesg mesg, G G, h h, Tr Tr, ntr ntr) Calculates and prints the global inventory of all tracers in the registry.
- Parameters
[in] mesg: message that appears on the chksum lines[in] g: ocean grid structure[in] tr: array of all of registered tracers[in] h: Layer thicknesses[in] ntr: number of registered tracers
-
subroutine, public mom_tracer_registry::tracer_name_lookup(Reg Reg, tr_ptr tr_ptr, name name) Find a tracer in the tracer registry by name.
- Parameters
reg: pointer to tracer registrytr_ptr: target or pointer to the tracer array[in] name: tracer name
-
subroutine, public mom_tracer_registry::tracer_registry_init(param_file param_file, Reg Reg) Initialize the tracer registry.
- Parameters
[in] param_file: open file to parse for model parametersreg: pointer to tracer registry
-
subroutine, public mom_tracer_registry::tracer_registry_end(Reg Reg) This routine closes the tracer registry module.
- Parameters
reg: The tracer registry that will be deallocated
-
-
namespace
mom_tracer_z_init¶ Used to initialize tracers from a depth- (or z*-) space file.
Functions
-
logical function, public mom_tracer_z_init::tracer_z_init(tr tr, h h, filename filename, tr_name tr_name, G G, US US, missing_val missing_val, land_val land_val) This function initializes a tracer by reading a Z-space file, returning .true. if this appears to have been successful, and false otherwise.
- Return
A return code indicating if the initialization has been successful
- Parameters
[in] g: The ocean’s grid structure[in] us: A dimensional unit scaling type[out] tr: The tracer to initialize[in] h: Layer thicknesses [H ~> m or kg m-2][in] filename: The name of the file to read from[in] tr_name: The name of the tracer in the file[in] missing_val: The missing value for the tracer[in] land_val: A value to use to fill in land points
-
subroutine
read_z_edges(filename filename, tr_name tr_name, z_edges z_edges, nz_out nz_out, has_edges has_edges, use_missing use_missing, missing missing, scale scale)¶ This subroutine reads the vertical coordinate data for a field from a NetCDF file. It also might read the missing value attribute for that same field.
- Parameters
[in] filename: The name of the file to read from.[in] tr_name: The name of the tracer in the file.[out] z_edges: The depths of the vertical edges of the tracer array[out] nz_out: The number of vertical layers in the tracer array[out] has_edges: If true the values in z_edges are the edges of the tracer cells, otherwise they are the cell centers[inout] use_missing: If false on input, see whether the tracer has a missing value, and if so return true[inout] missing: The missing value, if one has been found[in] scale: A scaling factor for z_edges into new units.
-
subroutine
find_overlap(e e, Z_top Z_top, Z_bot Z_bot, k_max k_max, k_start k_start, k_top k_top, k_bot k_bot, wt wt, z1 z1, z2 z2)¶ Determines the layers bounded by interfaces e that overlap with the depth range between Z_top and Z_bot, and the fractional weights of each layer. It also calculates the normalized relative depths of the range of each layer that overlaps that depth range.
- Parameters
[in] e: Column interface heights, in arbitrary units.[in] z_top: Top of range being mapped to, in the units of e.[in] z_bot: Bottom of range being mapped to, in the units of e.[in] k_max: Number of valid layers.[in] k_start: Layer at which to start searching.[inout] k_top: Indices of top layers that overlap with the depth range.[inout] k_bot: Indices of bottom layers that overlap with the depth range.[out] wt: Relative weights of each layer from k_top to k_bot.[out] z1: Depth of the top limits of the part of a layer that contributes to a depth level, relative to the cell center and normalized by the cell thickness [nondim]. Note that -1/2 <= z1 < z2 <= 1/2.[out] z2: Depths of the bottom limit of the part of a layer that contributes to a depth level, relative to the cell center and normalized by the cell thickness [nondim]. Note that -1/2 <= z1 < z2 <= 1/2.
-
subroutine
find_limited_slope(val val, e e, slope slope, k k)¶ This subroutine determines a limited slope for val to be advected with a piecewise limited scheme.
- Parameters
[in] val: A column of values that are being interpolated.[in] e: Column interface heights in arbitrary units[out] slope: Normalized slope in the intracell distribution of val.[in] k: Layer whose slope is being determined.
-
-
namespace
mom_transcribe_grid¶ Module with routines for copying information from a shared dynamic horizontal grid to an ocean-specific horizontal grid and the reverse.
Functions
-
subroutine, public mom_transcribe_grid::copy_dyngrid_to_mom_grid(dG dG, oG oG) Copies information from a dynamic (shared) horizontal grid type into an ocean_grid_type.
- Parameters
[in] dg: Common horizontal grid type[inout] og: Ocean grid type
-
subroutine, public mom_transcribe_grid::copy_mom_grid_to_dyngrid(oG oG, dG dG) Copies information from an ocean_grid_type into a dynamic (shared) horizontal grid type.
- Parameters
[in] og: Ocean grid type[inout] dg: Common horizontal grid type
-
-
namespace
mom_unit_scaling¶ Provides a transparent unit rescaling type to facilitate dimensional consistency testing.
Functions
-
subroutine, public mom_unit_scaling::unit_scaling_init(param_file param_file, US US) Allocates and initializes the ocean model unit scaling type.
- Parameters
[in] param_file: Parameter file handle/typeus: A dimensional unit scaling type
-
subroutine, public mom_unit_scaling::fix_restart_unit_scaling(US US) Set the unit scaling factors for output to restart files to the unit scaling factors for this run.
- Parameters
[inout] us: A dimensional unit scaling type
-
subroutine, public mom_unit_scaling::unit_scaling_end(US US) Deallocates a unit scaling structure.
- Parameters
us: A dimensional unit scaling type
-
-
namespace
mom_unit_tests¶ Invokes unit tests in all modules that have them.
Functions
-
subroutine, public mom_unit_tests::unit_tests(verbosity verbosity) Calls unit tests for other modules. Note that if a unit test returns true, a FATAL error is triggered.
- Parameters
[in] verbosity: The verbosity level
-
-
namespace
mom_variables¶ Provides transparent structures with groups of MOM6 variables and supporting routines.
Functions
-
subroutine, public mom_variables::allocate_surface_state(sfc_state sfc_state, G G, use_temperature use_temperature, do_integrals do_integrals, gas_fields_ocn gas_fields_ocn, use_meltpot use_meltpot) Allocates the fields for the surface (return) properties of the ocean model. Unused fields are unallocated.
- Parameters
[in] g: ocean grid structure[inout] sfc_state: ocean surface state type to be allocated.[in] use_temperature: If true, allocate the space for thermodynamic variables.[in] do_integrals: If true, allocate the space for vertically integrated fields.[in] gas_fields_ocn: If present, this type describes the ocean[in] use_meltpot: If true, allocate the space for melt potential
-
subroutine, public mom_variables::deallocate_surface_state(sfc_state sfc_state) Deallocates the elements of a surface state type.
- Parameters
[inout] sfc_state: ocean surface state type to be deallocated here.
-
subroutine, public mom_variables::alloc_bt_cont_type(BT_cont BT_cont, G G, alloc_faces alloc_faces) Allocates the arrays contained within a BT_cont_type and initializes them to 0.
- Parameters
bt_cont: The BT_cont_type whose elements will be allocated[in] g: The ocean’s grid structure[in] alloc_faces: If present and true, allocate memory for effective face thicknesses.
-
subroutine, public mom_variables::dealloc_bt_cont_type(BT_cont BT_cont) Deallocates the arrays contained within a BT_cont_type.
- Parameters
bt_cont: The BT_cont_type whose elements will be deallocated.
-
subroutine, public mom_variables::mom_thermovar_chksum(mesg mesg, tv tv, G G) Diagnostic checksums on various elements of a thermo_var_ptrs type for debugging.
- Parameters
[in] mesg: A message that appears in the checksum lines[in] tv: A structure pointing to various thermodynamic variables[in] g: The ocean’s grid structure
-
-
namespace
mom_vert_friction¶ Implements vertical viscosity (vertvisc)
The vertical diffusion of momentum is fully implicit. This is necessary to allow for vanishingly small layers. The coupling is based on the distance between the centers of adjacent layers, except where a layer is close to the bottom compared with a bottom boundary layer thickness when a bottom drag law is used. A stress top b.c. and a no slip bottom b.c. are used. There is no limit on the time step for vertvisc.
- Author
Robert Hallberg
- Date
April 1994 - October 2006
Near the bottom, the horizontal thickness interpolation scheme changes to an upwind biased estimate to control the effect of spurious Montgomery potential gradients at the bottom where nearly massless layers layers ride over the topography. Within a few boundary layer depths of the bottom, the harmonic mean thickness (i.e. (2 h+ h-) / (h+ + h-) ) is used if the velocity is from the thinner side and the arithmetic mean thickness (i.e. (h+ + h-)/2) is used if the velocity is from the thicker side. Both of these thickness estimates are second order accurate. Above this the arithmetic mean thickness is used.
In addition, vertvisc truncates any velocity component that exceeds maxvel to truncvel. This basically keeps instabilities spatially localized. The number of times the velocity is truncated is reported each time the energies are saved, and if exceeds CSMaxtrunc the model will stop itself and change the time to a large value. This has proven very useful in (1) diagnosing model failures and (2) letting the model settle down to a meaningful integration from a poorly specified initial condition.
The same code is used for the two velocity components, by indirectly referencing the velocities and defining a handful of direction-specific defined variables.
Macros written all in capital letters are defined in MOM_memory.h.
A small fragment of the grid is shown below:
j+1 x ^ x ^ x At x: q j+1 > o > o > At ^: v, frhatv, tauy j x ^ x ^ x At >: u, frhatu, taux j > o > o > At o: h j-1 x ^ x ^ x i-1 i i+1 At x & ^: i i+1 At > & o:
The boundaries always run through q grid points (x).
Functions
-
subroutine, public mom_vert_friction::vertvisc(u u, v v, h h, forces forces, visc visc, dt dt, OBC OBC, ADp ADp, CDp CDp, G G, GV GV, US US, CS CS, taux_bot taux_bot, tauy_bot tauy_bot, Waves Waves) Perform a fully implicit vertical diffusion of momentum. Stress top and bottom boundary conditions are used.
This is solving the tridiagonal system
\[ \left(h_k + a_{k + 1/2} + a_{k - 1/2} + r_k\right) u_k^{n+1} = h_k u_k^n + a_{k + 1/2} u_{k+1}^{n+1} + a_{k - 1/2} u_{k-1}^{n+1} \]where \(a_{k + 1/2} = \Delta t \nu_{k + 1/2} / h_{k + 1/2}\) is the interfacial coupling thickness per time step, encompassing background viscosity as well as contributions from enhanced mixed and bottom layer viscosities. $r_k$ is a Rayleight drag term due to channel drag. There is an additional stress term on the right-hand side if DIRECT_STRESS is true, applied to the surface layer.- Parameters
[in] g: Ocean grid structure[in] gv: Ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] u: Zonal velocity [m s-1][inout] v: Meridional velocity [m s-1][in] h: Layer thickness [H ~> m or kg m-2][in] forces: A structure with the driving mechanical forces[inout] visc: Viscosities and bottom drag[in] dt: Time increment [s]obc: Open boundary condition structure[inout] adp: Accelerations in the momentum equations for diagnostics[inout] cdp: Continuity equation termscs: Vertical viscosity control structure[out] taux_bot: Zonal bottom stress from ocean to rock [kg Z s-2 m-2 ~> Pa][out] tauy_bot: Meridional bottom stress from ocean to rock [kg Z s-2 m-2 ~> Pa]waves: Container for wave/Stokes information
-
subroutine, public mom_vert_friction::vertvisc_remnant(visc visc, visc_rem_u visc_rem_u, visc_rem_v visc_rem_v, dt dt, G G, GV GV, US US, CS CS) Calculate the fraction of momentum originally in a layer that remains after a time-step of viscosity, and the fraction of a time-step’s worth of barotropic acceleration that a layer experiences after viscosity is applied.
- Parameters
[in] g: Ocean grid structure[in] gv: Ocean vertical grid structure[in] visc: Viscosities and bottom drag[inout] visc_rem_u: Fraction of a time-step’s worth of a[inout] visc_rem_v: Fraction of a time-step’s worth of a[in] dt: Time increment [s][in] us: A dimensional unit scaling typecs: Vertical viscosity control structure
-
subroutine, public mom_vert_friction::vertvisc_coef(u u, v v, h h, forces forces, visc visc, dt dt, G G, GV GV, US US, CS CS, OBC OBC) Calculate the coupling coefficients (CSa_u and CSa_v) and effective layer thicknesses (CSh_u and CSh_v) for later use in the applying the implicit vertical viscosity via vertvisc().
- Parameters
[in] g: Ocean grid structure[in] gv: Ocean vertical grid structure[in] us: A dimensional unit scaling type[in] u: Zonal velocity [m s-1][in] v: Meridional velocity [m s-1][in] h: Layer thickness [H ~> m or kg m-2][in] forces: A structure with the driving mechanical forces[in] visc: Viscosities and bottom drag[in] dt: Time increment [s]cs: Vertical viscosity control structureobc: Open boundary condition structure
-
subroutine
find_coupling_coef(a_cpl a_cpl, hvel hvel, do_i do_i, h_harm h_harm, bbl_thick bbl_thick, kv_bbl kv_bbl, z_i z_i, h_ml h_ml, dt dt, j j, G G, GV GV, US US, CS CS, visc visc, forces forces, work_on_u work_on_u, OBC OBC, shelf shelf)¶ Calculate the ‘coupling coefficient’ (a_cpl) at the interfaces. If BOTTOMDRAGLAW is defined, the minimum of Hbbl and half the adjacent layer thicknesses are used to calculate a_cpl near the bottom.
- Parameters
[in] g: Ocean grid structure[in] gv: Ocean vertical grid structure[in] us: A dimensional unit scaling type[out] a_cpl: Coupling coefficient across interfaces [Z T-1 ~> m s-1].[in] hvel: Thickness at velocity points [H ~> m or kg m-2][in] do_i: If true, determine coupling coefficient for a column[in] h_harm: Harmonic mean of thicknesses around a velocity[in] bbl_thick: Bottom boundary layer thickness [H ~> m or kg m-2][in] kv_bbl: Bottom boundary layer viscosity [Z2 T-1 ~> m2 s-1].[in] z_i: Estimate of interface heights above the bottom,[out] h_ml: Mixed layer depth [H ~> m or kg m-2][in] j: j-index to find coupling coefficient for[in] dt: Time increment [s]cs: Vertical viscosity control structure[in] visc: Structure containing viscosities and bottom drag[in] forces: A structure with the driving mechanical forces[in] work_on_u: If true, u-points are being calculated, otherwise they are v-pointsobc: Open boundary condition structure[in] shelf: If present and true, use a surface boundary condition appropriate for an ice shelf.
-
subroutine, public mom_vert_friction::vertvisc_limit_vel(u u, v v, h h, ADp ADp, CDp CDp, forces forces, visc visc, dt dt, G G, GV GV, US US, CS CS) Velocity components which exceed a threshold for physically reasonable values are truncated. Optionally, any column with excessive velocities may be sent to a diagnostic reporting subroutine.
- Parameters
[in] g: Ocean grid structure[in] gv: Ocean vertical grid structure[in] us: A dimensional unit scaling type[inout] u: Zonal velocity [m s-1][inout] v: Meridional velocity [m s-1][in] h: Layer thickness [H ~> m or kg m-2][in] adp: Acceleration diagnostic pointers[in] cdp: Continuity diagnostic pointers[in] forces: A structure with the driving mechanical forces[in] visc: Viscosities and bottom drag[in] dt: Time increment [s]cs: Vertical viscosity control structure
-
subroutine, public mom_vert_friction::vertvisc_init(MIS MIS, Time Time, G G, GV GV, US US, param_file param_file, diag diag, ADp ADp, dirs dirs, ntrunc ntrunc, CS CS) Initialize the vertical friction module.
- Parameters
[in] mis: The “MOM Internal State”, a set of pointers[in] time: Current model time[in] g: Ocean grid structure[in] gv: Ocean vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: File to parse for parameters[inout] diag: Diagnostic control structure[inout] adp: Acceleration diagnostic pointers[in] dirs: Relevant directory paths[inout] ntrunc: Number of velocity truncationscs: Vertical viscosity control structure
-
subroutine, public mom_vert_friction::updatecfltruncationvalue(Time Time, CS CS, activate activate) Update the CFL truncation value as a function of time. If called with the optional argument activate=.true., record the value of Time as the beginning of the ramp period.
- Parameters
[in] time: Current model timecs: Vertical viscosity control structure[in] activate: Specifiy whether to record the value of Time as the beginning of the ramp period
-
subroutine, public mom_vert_friction::vertvisc_end(CS CS) Clean up and deallocate the vertical friction module.
- Parameters
cs: Vertical viscosity control structure that will be deallocated in this subroutine.
-
namespace
mom_verticalgrid¶ Provides a transparent vertical ocean grid type and supporting routines.
Functions
-
subroutine, public mom_verticalgrid::verticalgridinit(param_file param_file, GV GV, US US) Allocates and initializes the ocean model vertical grid structure.
- Parameters
[in] param_file: Parameter file handle/typegv: The container for vertical grid data[in] us: A dimensional unit scaling type
-
subroutine, public mom_verticalgrid::fix_restart_scaling(GV GV) Set the scaling factors for restart files to the scaling factors for this run.
- Parameters
[inout] gv: The ocean’s vertical grid structure
-
character(len=48) function, public mom_verticalgrid::get_thickness_units(GV GV) Returns the model’s thickness units, usually m or kg/m^2.
- Return
The vertical thickness units
- Parameters
[in] gv: The ocean’s vertical grid structure
-
character(len=48) function, public mom_verticalgrid::get_flux_units(GV GV) Returns the model’s thickness flux units, usually m^3/s or kg/s.
- Return
The thickness flux units
- Parameters
[in] gv: The ocean’s vertical grid structure
-
character(len=48) function, public mom_verticalgrid::get_tr_flux_units(GV GV, tr_units tr_units, tr_vol_conc_units tr_vol_conc_units, tr_mass_conc_units tr_mass_conc_units) Returns the model’s tracer flux units.
- Return
The model’s flux units for a tracer.
- Parameters
[in] gv: The ocean’s vertical grid structure.[in] tr_units: Units for a tracer, for example Celsius or PSU.[in] tr_vol_conc_units: The concentration units per unit volume, for example if the units are umol m-3, tr_vol_conc_units would be umol.[in] tr_mass_conc_units: The concentration units per unit mass of sea water, for example if the units are mol kg-1, tr_vol_conc_units would be mol.
-
subroutine, public mom_verticalgrid::setverticalgridaxes(Rlay Rlay, GV GV) This sets the coordinate data for the “layer mode” of the isopycnal model.
- Parameters
[inout] gv: The container for vertical grid data[in] rlay: The layer target density
-
subroutine, public mom_verticalgrid::verticalgridend(GV GV) Deallocates the model’s vertical grid structure.
- Parameters
gv: The ocean’s vertical grid structure
-
-
namespace
mom_wave_interface¶ Interface for surface waves.
This module should be moved as wave coupling progresses and likely will should mirror the iceberg or sea-ice model set-up.
- Author
Brandon Reichl, 2018.
This module is meant to contain the routines to read in and interpret surface wave data for MOM6. In its original form, the capabilities include setting the Stokes drift in the model (from a variety of sources including prescribed, empirical, and input files). In short order, the plan is to also ammend the subroutine to accept Stokes drift information from an external coupler. Eventually, it will be necessary to break this file apart so that general wave information may be stored in the control structure and the Stokes drift effect can be isolated from processes such as sea-state dependent momentum fluxes, gas fluxes, and other wave related air-sea interaction and boundary layer phenomenon.
The Stokes drift are stored on the C-grid with the conventional protocol to interpolate to the h-grid to compute Langmuir number, the primary quantity needed for Langmuir turbulence parameterizations in both the ePBL and KPP approach. This module also computes full 3d Stokes drift profiles, which will be useful if second-order type boundary layer parameterizations are implemented (perhaps via GOTM, work in progress).
Unnamed Group
-
integer
wavemethod= -99¶ Options for including wave information Valid (tested) choices are: 0 - Test Profile 1 - Surface Stokes Drift Bands 2 - DHH85 3 - LF17 -99 - No waves computed, but empirical Langmuir number used.
-
integer, public mom_wave_interface::numbands =0 Number of wavenumber/frequency partitions to receive This needs to match the number of bands provided via either coupling or file.
-
integer, public mom_wave_interface::partitionmode Method for partition mode (meant to check input) 0 - wavenumbers 1 - frequencies.
-
integer
datasource¶ Integer that specifies where the Model Looks for Data Valid choices are: 1 - FMS DataOverride Routine 2 - Reserved For Coupler 3 - User input (fixed values, useful for 1d testing)
-
character(len=40) mom_wave_interface::surfbandfilename Filename if using DataOverride.
-
logical
dataoverrideisinitialized¶ Flag for DataOverride Initialization.
-
real
la_frachbl¶ Fraction of OSBL for averaging Langmuir number.
-
logical
la_misalignment= .false.¶ Flag to use misalignment in Langmuir number.
-
character(len=40) mom_wave_interface::mdl = "MOM_wave_interface" This module’s name.
-
integer, parameter mom_wave_interface::testprof = 0 Undocumented parameters.
-
integer, parameter mom_wave_interface::surfbands = 1 Options for including wave information Valid (tested) choices are: 0 - Test Profile 1 - Surface Stokes Drift Bands 2 - DHH85 3 - LF17 -99 - No waves computed, but empirical Langmuir number used.
-
integer, parameter mom_wave_interface::dhh85 = 2 Options for including wave information Valid (tested) choices are: 0 - Test Profile 1 - Surface Stokes Drift Bands 2 - DHH85 3 - LF17 -99 - No waves computed, but empirical Langmuir number used.
-
integer, parameter mom_wave_interface::lf17 = 3 Options for including wave information Valid (tested) choices are: 0 - Test Profile 1 - Surface Stokes Drift Bands 2 - DHH85 3 - LF17 -99 - No waves computed, but empirical Langmuir number used.
-
integer, parameter mom_wave_interface::null_wavemethod =-99 Options for including wave information Valid (tested) choices are: 0 - Test Profile 1 - Surface Stokes Drift Bands 2 - DHH85 3 - LF17 -99 - No waves computed, but empirical Langmuir number used.
-
integer, parameter mom_wave_interface::dataovr = 1 Options for including wave information Valid (tested) choices are: 0 - Test Profile 1 - Surface Stokes Drift Bands 2 - DHH85 3 - LF17 -99 - No waves computed, but empirical Langmuir number used.
-
integer, parameter mom_wave_interface::coupler = 2 Options for including wave information Valid (tested) choices are: 0 - Test Profile 1 - Surface Stokes Drift Bands 2 - DHH85 3 - LF17 -99 - No waves computed, but empirical Langmuir number used.
-
integer, parameter mom_wave_interface::input = 3 Options for including wave information Valid (tested) choices are: 0 - Test Profile 1 - Surface Stokes Drift Bands 2 - DHH85 3 - LF17 -99 - No waves computed, but empirical Langmuir number used.
-
real
tp_stkx0¶ Options for including wave information Valid (tested) choices are: 0 - Test Profile 1 - Surface Stokes Drift Bands 2 - DHH85 3 - LF17 -99 - No waves computed, but empirical Langmuir number used.
-
real
tp_stky0¶ Options for including wave information Valid (tested) choices are: 0 - Test Profile 1 - Surface Stokes Drift Bands 2 - DHH85 3 - LF17 -99 - No waves computed, but empirical Langmuir number used.
-
real
tp_wvl¶ Options for including wave information Valid (tested) choices are: 0 - Test Profile 1 - Surface Stokes Drift Bands 2 - DHH85 3 - LF17 -99 - No waves computed, but empirical Langmuir number used.
-
logical
waveagepeakfreq¶ Options for including wave information Valid (tested) choices are: 0 - Test Profile 1 - Surface Stokes Drift Bands 2 - DHH85 3 - LF17 -99 - No waves computed, but empirical Langmuir number used.
-
logical
staticwaves¶ Options for including wave information Valid (tested) choices are: 0 - Test Profile 1 - Surface Stokes Drift Bands 2 - DHH85 3 - LF17 -99 - No waves computed, but empirical Langmuir number used.
-
logical
dhh85_is_set¶ Options for including wave information Valid (tested) choices are: 0 - Test Profile 1 - Surface Stokes Drift Bands 2 - DHH85 3 - LF17 -99 - No waves computed, but empirical Langmuir number used.
-
real
waveage¶ Options for including wave information Valid (tested) choices are: 0 - Test Profile 1 - Surface Stokes Drift Bands 2 - DHH85 3 - LF17 -99 - No waves computed, but empirical Langmuir number used.
-
real
wavewind¶ Options for including wave information Valid (tested) choices are: 0 - Test Profile 1 - Surface Stokes Drift Bands 2 - DHH85 3 - LF17 -99 - No waves computed, but empirical Langmuir number used.
-
real
pi¶ Options for including wave information Valid (tested) choices are: 0 - Test Profile 1 - Surface Stokes Drift Bands 2 - DHH85 3 - LF17 -99 - No waves computed, but empirical Langmuir number used.
-
subroutine, public mom_wave_interface::mom_wave_interface_init(time time, G G, GV GV, US US, param_file param_file, CS CS, diag diag) Initializes parameters related to MOM_wave_interface.
- Parameters
[in] time: Model time[inout] g: Grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: Input parameter structurecs: Wave parameter control structure[inout] diag: Diagnostic Pointer
-
subroutine, public mom_wave_interface::mom_wave_interface_init_lite(param_file param_file) A ‘lite’ init subroutine to initialize a few inputs needed if using wave information with the wind-speed dependent Stokes drift formulation of LF17.
- Parameters
[in] param_file: Input parameter structure
-
subroutine, public mom_wave_interface::update_surface_waves(G G, GV GV, US US, Day Day, dt dt, CS CS) Subroutine that handles updating of surface wave/Stokes drift related properties.
- Parameters
cs: Wave parameter Control structure[inout] g: Grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[in] day: Current model time[in] dt: Timestep as a time-type
-
subroutine, public mom_wave_interface::update_stokes_drift(G G, GV GV, US US, CS CS, h h, ustar ustar) Constructs the Stokes Drift profile on the model grid based on desired coupling options.
- Parameters
cs: Wave parameter Control structure[inout] g: Grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[in] h: Thickness [H ~> m or kg m-2][in] ustar: Wind friction velocity [Z T-1 ~> m s-1].
-
subroutine
surface_bands_by_data_override(day_center day_center, G G, GV GV, US US, CS CS)¶ A subroutine to fill the Stokes drift from a NetCDF file using the data_override procedures.
- Parameters
[in] day_center: Center of timestepcs: Wave structure[inout] g: Grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type
-
subroutine
get_stokessl_lifoxkemper(ustar ustar, hbl hbl, GV GV, US US, UStokes_SL UStokes_SL, LA LA)¶ Get SL averaged Stokes drift from Li/FK 17 method.
Original description:
This function returns the enhancement factor, given the 10-meter wind [m s-1], friction velocity [m s-1] and the boundary layer depth [m].
Update (Jan/25):
Converted from function to subroutine, now returns Langmuir number.
Computs 10m wind internally, so only ustar and hbl need passed to subroutine.
Qing Li, 160606
BGR port from CVMix to MOM6 Jan/25/2017
BGR change output to LA from Efactor
BGR remove u10 input
BGR note: fixed parameter values should be changed to “get_params”
- Parameters
[in] ustar: water-side surface friction velocity [Z T-1 ~> m s-1].[in] hbl: boundary layer depth [Z ~> m].[in] gv: Ocean vertical grid structure[in] us: A dimensional unit scaling type[out] ustokes_sl: Surface layer averaged Stokes drift [m s-1][out] la: Langmuir number
-
subroutine
get_sl_average_prof(GV GV, AvgDepth AvgDepth, H H, Profile Profile, Average Average)¶ Get SL Averaged Stokes drift from a Stokes drift Profile.
- Parameters
[in] gv: Ocean vertical grid structure[in] avgdepth: Depth to average over [Z ~> m].[in] h: Grid thickness [H ~> m or kg m-2][in] profile: Profile of quantity to be averaged [arbitrary][out] average: Output quantity averaged over depth AvgDepth [arbitrary] (used here for Stokes drift)
-
subroutine
get_sl_average_band(GV GV, AvgDepth AvgDepth, NB NB, WaveNumbers WaveNumbers, SurfStokes SurfStokes, Average Average)¶ Get SL averaged Stokes drift from the banded Spectrum method.
- Parameters
[in] gv: Ocean vertical grid[in] avgdepth: Depth to average over [Z ~> m].[in] nb: Number of bands used[in] wavenumbers: Wavenumber corresponding to each band [Z-1 ~> m-1][in] surfstokes: Surface Stokes drift for each band [m s-1][out] average: Output average Stokes drift over depth AvgDepth [m s-1]
-
subroutine
dhh85_mid(GV GV, US US, zpt zpt, UStokes UStokes)¶ Compute the Stokes drift at a given depth.
Taken from Qing Li (Brown) use for comparing MOM6 simulation to his LES computed at z mid point (I think) and not depth averaged. Should be fine to integrate in frequency from 0.1 to sqrt(-0.2*grav*2pi/dz
- Parameters
[in] gv: Ocean vertical grid[in] us: A dimensional unit scaling type[in] zpt: Depth to get Stokes drift [Z ~> m]. !### THIS IS NOT USED YET.[out] ustokes: Stokes drift [m s-1]
-
subroutine, public mom_wave_interface::stokesmixing(G G, GV GV, dt dt, h h, u u, v v, Waves Waves) Explicit solver for Stokes mixing. Still in development do not use.
- Parameters
[in] g: Ocean grid[in] gv: Ocean vertical grid[in] dt: Time step of MOM6 [T ~> s] for explicit solver[in] h: Layer thicknesses [H ~> m or kg m-2][inout] u: Velocity i-component [m s-1][inout] v: Velocity j-component [m s-1]waves: Surface wave related control structure.
-
subroutine, public mom_wave_interface::coriolisstokes(G G, GV GV, DT DT, h h, u u, v v, WAVES WAVES, US US) Solver to add Coriolis-Stokes to model Still in development and not meant for general use. Can be activated (with code intervention) for LES comparison CHECK THAT RIGHT TIMESTEP IS PASSED IF YOU USE THIS**.
Not accessed in the standard code.
- Parameters
[in] g: Ocean grid[in] gv: Ocean vertical grid[in] dt: Time step of MOM6 [s] CHECK IF PASSING RIGHT TIMESTEP[in] h: Layer thicknesses [H ~> m or kg m-2][inout] u: Velocity i-component [m s-1][inout] v: Velocity j-component [m s-1]waves: Surface wave related control structure.[in] us: A dimensional unit scaling type
-
subroutine
ust_2_u10_coare3p5(USTair USTair, U10 U10, GV GV, US US)¶ Computes wind speed from ustar_air based on COARE 3.5 Cd relationship Probably doesn’t belong in this module, but it is used here to estimate wind speed for wind-wave relationships. Should be a fine way to estimate the neutral wind-speed as written here.
- Parameters
[in] ustair: Wind friction velocity [m s-1][out] u10: 10-m neutral wind speed [m s-1][in] gv: vertical grid type[in] us: A dimensional unit scaling type
-
subroutine, public mom_wave_interface::waves_end(CS CS) Clear pointers, deallocate memory.
- Parameters
cs: Control structure
Functions
-
subroutine, public mom_wave_interface::get_langmuir_number(LA LA, G G, GV GV, US US, HBL HBL, ustar ustar, i i, j j, H H, U_H U_H, V_H V_H, Override_MA Override_MA, Waves Waves) Interface to get Langmuir number based on options stored in wave structure.
Note this can be called with an unallocated Waves pointer, which is okay if we want the wind-speed only dependent Langmuir number. Therefore, we need to be careful about what we try to access here.
- Parameters
[in] g: Ocean grid structure[in] gv: Ocean vertical grid structure[in] us: A dimensional unit scaling type[in] i: Meridional index of h-point[in] j: Zonal index of h-point[in] ustar: Friction velocity [Z T-1 ~> m s-1].[in] hbl: (Positive) thickness of boundary layer [Z ~> m].[in] override_ma: Override to use misalignment in LA calculation. This can be used if diagnostic LA outputs are desired that are different than those used by the dynamical model.[in] h: Grid layer thickness [H ~> m or kg m-2][in] u_h: Zonal velocity at H point [m s-1][in] v_h: Meridional velocity at H point [m s-1]waves: Surface wave control structure.[out] la: Langmuir number
-
namespace
mom_wave_speed¶ Routines for calculating baroclinic wave speeds.
Subroutine wave_speed() solves for the first baroclinic mode wave speed. (It could solve for all the wave speeds, but the iterative approach taken here means that this is not particularly efficient.)
If
e(k)is the perturbation interface height, this means solving for the smallest eigenvalue (lam= 1/c^2) of the system-Igu(k)*e(k-1) + (Igu(k)+Igl(k)-lam)*e(k) - Igl(k)*e(k+1) = 0.0
with rigid lid boundary conditions e(1) = e(nz+1) = 0.0 giving
(Igu(2)+Igl(2)-lam)*e(2) - Igl(2)*e(3) = 0.0 -Igu(nz)*e(nz-1) + (Igu(nz)+Igl(nz)-lam)*e(nz) = 0.0
Here
Igl(k) = 1.0/(gprime(k)*h(k)) ; Igu(k) = 1.0/(gprime(k)*h(k-1))
Alternately, these same eigenvalues can be found from the second smallest eigenvalue of the Montgomery potential (M(k)) calculation:
-Igl(k)*M(k-1) + (Igl(k)+Igu(k+1)-lam)*M(k) - Igu(k+1)*M(k+1) = 0.0
with rigid lid and flat bottom boundary conditions
(Igu(2)-lam)*M(1) - Igu(2)*M(2) = 0.0 -Igl(nz)*M(nz-1) + (Igl(nz)-lam)*M(nz) = 0.0
Note that the barotropic mode has been eliminated from the rigid lid interface height equations, hence the matrix is one row smaller. Without the rigid lid, the top boundary condition is simpler to implement with the M equations.
Functions
-
subroutine, public mom_wave_speed::wave_speed(h h, tv tv, G G, GV GV, US US, cg1 cg1, CS CS, full_halos full_halos, use_ebt_mode use_ebt_mode, mono_N2_column_fraction mono_N2_column_fraction, mono_N2_depth mono_N2_depth, modal_structure modal_structure) Calculates the wave speed of the first baroclinic mode.
- Parameters
[in] g: Ocean grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[in] h: Layer thickness [H ~> m or kg m-2][in] tv: Thermodynamic variables[out] cg1: First mode internal wave speed [m s-1]cs: Control structure for MOM_wave_speed[in] full_halos: If true, do the calculation over the entire computational domain.[in] use_ebt_mode: If true, use the equivalent barotropic mode instead of the first baroclinic mode.[in] mono_n2_column_fraction: The lower fraction of water column over which N2 is limited as monotonic for the purposes of calculating vertical modal structure.[in] mono_n2_depth: A depth below which N2 is limited as monotonic for the purposes of calculating vertical modal structure [m].[out] modal_structure: Normalized model structure [nondim]
-
subroutine
tdma6(n n, a a, b b, c c, lam lam, y y)¶ Solve a non-symmetric tridiagonal problem with a scalar contribution to the leading diagonal. This uses the Thomas algorithm rather than the Hallberg algorithm since the matrix is not symmetric.
- Parameters
[in] n: Number of rows of matrix[in] a: Lower diagonal[in] b: Leading diagonal[in] c: Upper diagonal[in] lam: Scalar subtracted from leading diagonal[inout] y: RHS on entry, result on exit
-
subroutine, public mom_wave_speed::wave_speeds(h h, tv tv, G G, GV GV, US US, nmodes nmodes, cn cn, CS CS, full_halos full_halos) Calculates the wave speeds for the first few barolinic modes.
- Parameters
[in] g: Ocean grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[in] h: Layer thickness [H ~> m or kg m-2][in] tv: Thermodynamic variables[in] nmodes: Number of modes[out] cn: Waves speeds [m s-1]cs: Control structure for MOM_wave_speed[in] full_halos: If true, do the calculation over the entire computational domain.
-
subroutine
tridiag_det(a a, b b, c c, nrows nrows, lam lam, det_out det_out, ddet_out ddet_out)¶ Calculate the determinant of a tridiagonal matrix with diagonals a,b-lam,c where lam is constant across rows.
- Parameters
[in] a: Lower diagonal of matrix (first entry = 0)[in] b: Leading diagonal of matrix (excluding lam)[in] c: Upper diagonal of matrix (last entry = 0)[in] nrows: Size of matrix[in] lam: Value subtracted from b[out] det_out: Determinant[out] ddet_out: Derivative of determinant w.r.t. lam
-
subroutine, public mom_wave_speed::wave_speed_init(CS CS, use_ebt_mode use_ebt_mode, mono_N2_column_fraction mono_N2_column_fraction, mono_N2_depth mono_N2_depth) Initialize control structure for MOM_wave_speed.
- Parameters
cs: Control structure for MOM_wave_speed[in] use_ebt_mode: If true, use the equivalent barotropic mode instead of the first baroclinic mode.[in] mono_n2_column_fraction: The lower fraction of water column over which N2 is limited as monotonic for the purposes of calculating the vertical modal structure.[in] mono_n2_depth: The depth below which N2 is limited as monotonic for the purposes of calculating the vertical modal structure.
-
subroutine, public mom_wave_speed::wave_speed_set_param(CS CS, use_ebt_mode use_ebt_mode, mono_N2_column_fraction mono_N2_column_fraction, mono_N2_depth mono_N2_depth) Sets internal parameters for MOM_wave_speed.
- Parameters
cs: Control structure for MOM_wave_speed[in] use_ebt_mode: If true, use the equivalent barotropic mode instead of the first baroclinic mode.[in] mono_n2_column_fraction: The lower fraction of water column over which N2 is limited as monotonic for the purposes of calculating the vertical modal structure.[in] mono_n2_depth: The depth below which N2 is limited as monotonic for the purposes of calculating the vertical modal structure.
-
-
namespace
mom_wave_structure¶ Vertical structure functions for first baroclinic mode wave speed.
Functions
-
subroutine, public mom_wave_structure::wave_structure(h h, tv tv, G G, GV GV, US US, cn cn, ModeNum ModeNum, freq freq, CS CS, En En, full_halos full_halos) This subroutine determines the internal wave velocity structure for any mode.
This subroutine solves for the eigen vector [vertical structure, e(k)] associated with the first baroclinic mode speed [i.e., smallest eigen value (lam = 1/c^2)] of the system d2e/dz2 = -(N2/cn2)e, or (A-lam*I)e = 0, where A = -(1/N2)(d2/dz2), lam = 1/c^2, and I is the identity matrix. 2nd order discretization in the vertical lets this system be represented as
-Igu(k)*e(k-1) + (Igu(k)+Igl(k)-lam)*e(k) - Igl(k)*e(k+1) = 0.0
with rigid lid boundary conditions e(1) = e(nz+1) = 0.0 giving
(Igu(2)+Igl(2)-lam)*e(2) - Igl(2)*e(3) = 0.0 -Igu(nz)*e(nz-1) + (Igu(nz)+Igl(nz)-lam)*e(nz) = 0.0
where, upon noting N2 = reduced gravity/layer thickness, we get Igl(k) = 1.0/(gprime(k)*H(k)) ; Igu(k) = 1.0/(gprime(k)*H(k-1))
The eigen value for this system is approximated using “wave_speed.” This subroutine uses these eigen values (mode speeds) to estimate the corresponding eigen vectors (velocity structure) using the “inverse iteration with shift” method. The algorithm is
Pick a starting vector reasonably close to mode structure and with unit magnitude, b_guess For n=1,2,3,… Solve (A-lam*I)e = e_guess for e Set e_guess=e/|e| and repeat, with each iteration refining the estimate of e
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[in] h: Layer thicknesses [H ~> m or kg m-2][in] tv: A structure pointing to various thermodynamic variables.[in] cn: The (non-rotational) mode internal gravity wave speed [m s-1].[in] modenum: Mode number[in] freq: Intrinsic wave frequency [s-1].cs: The control structure returned by a previous call to wave_structure_init.[in] en: Internal wave energy density [J m-2].[in] full_halos: If true, do the calculation over the entire computational domain.
-
subroutine
tridiag_solver(a a, b b, c c, h h, y y, method method, x x)¶ Solves a tri-diagonal system Ax=y using either the standard Thomas algorithm (TDMA_T) or its more stable variant that invokes the “Hallberg substitution” (TDMA_H).
- Parameters
[in] a: lower diagonal with first entry equal to zero.[in] b: middle diagonal.[in] c: upper diagonal with last entry equal to zero.[in] h: vector of values that have already been added to b; used for systems of the form (e.g. average layer thickness in vertical diffusion case): [ -alpha(k-1/2) ] * e(k-1) + [ alpha(k-1/2) + alpha(k+1/2) + h(k) ] * e(k) + [ -alpha(k+1/2) ] * e(k+1) = y(k) where a(k)=[-alpha(k-1/2)], b(k)=[alpha(k-1/2)+alpha(k+1/2) + h(k)], and c(k)=[-alpha(k+1/2)]. Only used with TDMA_H method.[in] y: vector of known values on right hand side.[in] method: A string describing the algorithm to use[out] x: vector of unknown values to solve for.
-
subroutine, public mom_wave_structure::wave_structure_init(Time Time, G G, param_file param_file, diag diag, CS CS) Allocate memory associated with the wave structure module and read parameters.
- Parameters
[in] time: The current model time.[in] g: The ocean’s grid structure.[in] param_file: A structure to parse for run-time parameters.[in] diag: A structure that is used to regulate diagnostic output.cs: A pointer that is set to point to the control structure for this module.
-
-
namespace
mom_write_cputime¶ A module to monitor the overall CPU time used by MOM6 and project when to stop the model.
By Robert Hallberg, May 2006.
This file contains the subroutine (write_cputime) that writes the summed CPU time across all processors to an output file. In addition, write_cputime estimates how many more time steps can be taken before 95% of the available CPU time is used, so that the model can be checkpointed at that time.
Functions
-
subroutine, public mom_write_cputime::write_cputime_start_clock(CS CS) Evaluate the CPU time returned by SYSTEM_CLOCK at the start of a run.
- Parameters
cs: The control structure set up by a previous call to MOM_write_cputime_init.
-
subroutine, public mom_write_cputime::mom_write_cputime_init(param_file param_file, directory directory, Input_start_time Input_start_time, CS CS) Initialize the MOM_write_cputime module.
- Parameters
[in] param_file: A structure to parse for run-time parameters[in] directory: The directory where the CPU time file goes.[in] input_start_time: The start model time of the simulation.cs: A pointer that may be set to point to the control structure for this module.
-
subroutine, public mom_write_cputime::write_cputime(day day, n n, nmax nmax, CS CS) This subroutine assesses how much CPU time the model has taken and determines how long the model should be run before it saves a restart file and stops itself.
- Parameters
[inout] day: The current model time.[in] n: The time step number of the current execution.[inout] nmax: The number of iterations after which to stop so that the simulation will not run out of CPU time.cs: The control structure set up by a previous call to MOM_write_cputime_init.
-
-
namespace
NETCDF¶
-
namespace
netcdf¶
-
namespace
neverland_initialization¶ Initialization for the “Neverland” configuration.
Functions
-
subroutine, public neverland_initialization::neverland_initialize_topography(D D, G G, param_file param_file, max_depth max_depth) This subroutine sets up the Neverland test case topography.
- Parameters
[in] g: The dynamic horizontal grid type[out] d: Ocean bottom depth in the units of depth_max[in] param_file: Parameter file structure[in] max_depth: Maximum ocean depth in arbitrary units
-
real function neverland_initialization::cosbell(x x, L L) Returns the value of a cosine-bell function evaluated at x/L.
- Parameters
[in] x: non-dimensional position[in] l: non-dimensional width
-
real function neverland_initialization::spike(x x, L L) Returns the value of a sin-spike function evaluated at x/L.
- Parameters
[in] x: non-dimensional position[in] l: non-dimensional width
-
subroutine, public neverland_initialization::neverland_initialize_thickness(h h, G G, GV GV, US US, param_file param_file, eqn_of_state eqn_of_state, P_ref P_ref) This subroutine initializes layer thicknesses for the Neverland test case, by finding the depths of interfaces in a specified latitude-dependent temperature profile with an exponentially decaying thermocline on top of a linear stratification.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[out] h: The thickness that is being initialized [H ~> m or kg m-2].[in] param_file: A structure indicating the open file to parse for model parameter values.eqn_of_state: integer that selects the equation of state.[in] p_ref: The coordinate-density reference pressure [Pa].
-
-
namespace
neverland_surface_forcing¶ Wind and buoyancy forcing for the Neverland configurations.
Functions
-
subroutine, public neverland_surface_forcing::neverland_wind_forcing(sfc_state sfc_state, forces forces, day day, G G, US US, CS CS) Sets the surface wind stresses, forcestaux and forcestauy for the Neverland forcing configuration.
- Parameters
[inout] sfc_state: A structure containing fields that describe the surface state of the ocean.[inout] forces: A structure with the driving mechanical forces[in] day: Time used for determining the fluxes.[inout] g: Grid structure.[in] us: A dimensional unit scaling typecs: Control structure for this module.
-
real function neverland_surface_forcing::cosbell(x x, L L) Returns the value of a cosine-bell function evaluated at x/L.
- Parameters
[in] x: non-dimensional position[in] l: non-dimensional width
-
real function neverland_surface_forcing::spike(x x, L L) Returns the value of a sin-spike function evaluated at x/L.
- Parameters
[in] x: non-dimensional position[in] l: non-dimensional width
-
subroutine, public neverland_surface_forcing::neverland_buoyancy_forcing(sfc_state sfc_state, fluxes fluxes, day day, dt dt, G G, US US, CS CS) Surface fluxes of buoyancy for the Neverland configurations.
- Parameters
[inout] sfc_state: A structure containing fields that describe the surface state of the ocean.[inout] fluxes: Forcing fields.[in] day: Time used for determining the fluxes.[in] dt: Forcing time step (s).[inout] g: Grid structure.[in] us: A dimensional unit scaling typecs: Control structure for this module.
-
subroutine, public neverland_surface_forcing::neverland_surface_forcing_init(Time Time, G G, US US, param_file param_file, diag diag, CS CS) Initializes the Neverland control structure.
- Parameters
[in] time: The current model time.[in] g: The ocean’s grid structure.[in] us: A dimensional unit scaling type[in] param_file: A structure indicating the open file to parse for model parameter values.[in] diag: A structure that is used to regulate diagnostic output.cs: A pointer that is set to point to the control structure for this module
-
-
namespace
ocean_model_mod¶ Top-level module for the MOM6 ocean model in coupled mode.
Functions
-
subroutine, public ocean_model_mod::ocean_model_init(Ocean_sfc Ocean_sfc, OS OS, Time_init Time_init, Time_in Time_in, gas_fields_ocn gas_fields_ocn) ocean_model_init initializes the ocean model, including registering fields for restarts and reading restart files if appropriate.
This subroutine initializes both the ocean state and the ocean surface type. Because of the way that indicies and domains are handled, Ocean_sfc must have been used in a previous call to initialize_ocean_type.
- Parameters
[inout] ocean_sfc: A structure containing various publiclyos: A structure whose internal contents are private to ocean_model_mod that may be used to contain all information about the ocean’s interior state.[in] time_init: The start time for the coupled model’s calendar[in] time_in: The time at which to initialize the ocean model.[in] gas_fields_ocn: If present, this type describes the
-
subroutine, public ocean_model_mod::update_ocean_model(Ice_ocean_boundary Ice_ocean_boundary, OS OS, Ocean_sfc Ocean_sfc, time_start_update time_start_update, Ocean_coupling_time_step Ocean_coupling_time_step, update_dyn update_dyn, update_thermo update_thermo, Ocn_fluxes_used Ocn_fluxes_used, start_cycle start_cycle, end_cycle end_cycle, cycle_length cycle_length) update_ocean_model uses the forcing in Ice_ocean_boundary to advance the ocean model’s state from the input value of Ocean_state (which must be for time time_start_update) for a time interval of Ocean_coupling_time_step, returning the publicly visible ocean surface properties in Ocean_sfc and storing the new ocean properties in Ocean_state.
- Parameters
[in] ice_ocean_boundary: A structure containing the variousos: A pointer to a private structure containing the[inout] ocean_sfc: A structure containing all the publicly visible[in] time_start_update: The time at the beginning of the update step.[in] ocean_coupling_time_step: The amount of time over which to advance the ocean.[in] update_dyn: If present and false, do not do updates due to the ocean dynamics.[in] update_thermo: If present and false, do not do updates due to the ocean thermodynamics or remapping.[in] ocn_fluxes_used: If present, this indicates whether the cumulative thermodynamic fluxes from the ocean, like frazil, have been used and should be reset.[in] start_cycle: This indicates whether this call is to be treated as the first call to step_MOM in a time-stepping cycle; missing is like true.[in] end_cycle: This indicates whether this call is to be treated as the last call to step_MOM in a time-stepping cycle; missing is like true.[in] cycle_length: The duration of a coupled time stepping cycle [s].
-
subroutine, public ocean_model_mod::ocean_model_restart(OS OS, timestamp timestamp) This subroutine writes out the ocean model restart file.
- Parameters
os: A pointer to the structure containing the internal ocean state being saved to a restart file[in] timestamp: An optional timestamp string that should be prepended to the file name. (Currently this is unused.)
-
subroutine, public ocean_model_mod::ocean_model_end(Ocean_sfc Ocean_sfc, Ocean_state Ocean_state, Time Time) ocean_model_end terminates the model run, saving the ocean state in a restart and deallocating any data associated with the ocean.
- Parameters
[inout] ocean_sfc: An ocean_public_type structure that is to be deallocated upon termination.ocean_state: A pointer to the structure containing the internal ocean state to be deallocated upon termination.[in] time: The model time, used for writing restarts.
-
subroutine, public ocean_model_mod::ocean_model_save_restart(OS OS, Time Time, directory directory, filename_suffix filename_suffix) ocean_model_save_restart causes restart files associated with the ocean to be written out.
- Parameters
os: A pointer to the structure containing the internal ocean state (in).[in] time: The model time at this call, needed for mpp_write calls.[in] directory: An optional directory into which to write these restart files.[in] filename_suffix: An optional suffix (e.g., a time-stamp) to append to the restart file names.
-
subroutine
initialize_ocean_public_type(input_domain input_domain, Ocean_sfc Ocean_sfc, diag diag, maskmap maskmap, gas_fields_ocn gas_fields_ocn)¶ Initialize the public ocean type.
- Parameters
[in] input_domain: The ocean model domain description[inout] ocean_sfc: A structure containing various publicly visible ocean surface properties after initialization, whose elements are allocated here.[in] diag: A structure that regulates diagnsotic output[in] maskmap: A mask indicating which virtual processors[in] gas_fields_ocn: If present, this type describes the
-
subroutine
convert_state_to_ocean_type(sfc_state sfc_state, Ocean_sfc Ocean_sfc, G G, patm patm, press_to_z press_to_z)¶ This subroutine translates the coupler’s ocean_data_type into MOM’s surface state variable. This may eventually be folded into the MOM code that calculates the surface state in the first place. Note the offset in the arrays because the ocean_data_type has no halo points in its arrays and always uses absolute indicies.
- Parameters
[inout] sfc_state: A structure containing fields that describe the surface state of the ocean.[inout] ocean_sfc: A structure containing various publicly[inout] g: The ocean’s grid structure[in] patm: The pressure at the ocean surface [Pa].[in] press_to_z: A conversion factor between pressure and ocean depth in m, usually 1/(rho_0*g) [m Pa-1].
-
subroutine, public ocean_model_mod::ocean_model_init_sfc(OS OS, Ocean_sfc Ocean_sfc) This subroutine extracts the surface properties from the ocean’s internal state and stores them in the ocean type returned to the calling ice model. It has to be separate from the ocean_initialization call because the coupler module allocates the space for some of these variables.
- Parameters
os: The structure with the complete ocean state[inout] ocean_sfc: A structure containing various publicly visible ocean surface properties after initialization, whose elements have their data set here.
-
subroutine, public ocean_model_mod::ocean_model_flux_init(OS OS, verbosity verbosity) ocean_model_flux_init is used to initialize properties of the air-sea fluxes as determined by various run-time parameters. It can be called from non-ocean PEs, or PEs that have not yet been initialzed, and it can safely be called multiple times.
- Parameters
os: An optional pointer to the ocean state, used to figure out if this is an ocean PE that has already been initialized.[in] verbosity: A 0-9 integer indicating a level of verbosity.
-
subroutine, public ocean_model_mod::ocean_stock_pe(OS OS, index index, value value, time_index time_index) Ocean_stock_pe - returns the integrated stocks of heat, water, etc. for conservation checks. Because of the way FMS is coded, only the root PE has the integrated amount, while all other PEs get 0.
- Parameters
os: A structure containing the internal ocean state. The data in OS is intent in.[in] index: The stock index for the quantity of interest.[out] value: Sum returned for the conservation quantity of interest.[in] time_index: An unused optional argument, present only for interfacial compatibility with other models.
-
subroutine
ocean_model_data2d_get(OS OS, Ocean Ocean, name name, array2D array2D, isc isc, jsc jsc)¶ This subroutine extracts a named 2-D field from the ocean surface or public type.
- Parameters
os: A pointer to the structure containing the internal ocean state (intent in).[in] ocean: A structure containing various publicly visible ocean surface fields.[in] name: The name of the field to extract[out] array2d: The values of the named field, it must cover only the computational domain[in] isc: The starting i-index of array2D[in] jsc: The starting j-index of array2D
-
subroutine
ocean_model_data1d_get(OS OS, Ocean Ocean, name name, value value)¶ This subroutine extracts a named scalar field from the ocean surface or public type.
- Parameters
os: A pointer to the structure containing the internal ocean state (intent in).[in] ocean: A structure containing various publicly visible ocean surface fields.[in] name: The name of the field to extract[out] value: The value of the named field
-
subroutine, public ocean_model_mod::ocean_public_type_chksum(id id, timestep timestep, ocn ocn) Write out FMS-format checsums on fields from the ocean surface state.
- Parameters
[in] id: An identifying string for this call[in] timestep: The number of elapsed timesteps[in] ocn: A structure containing various publicly visible ocean surface fields.
-
-
namespace
oil_tracer¶ A tracer package to mimic dissolved oil.
By Alistair Adcroft and Robert Hallberg, 2010 *
In the midst of the Deepwater Horizon oil spill, it became evident that models were needed to predict the long-term fate of dissolved oil in the open ocean. This tracer packages mimics the transport, dilution and decay of dissolved oil plumes in the ocean.
This tracer package was central to the simulations used by Adcroft et al., GRL 2010, to prove that the Deepwater Horizon spill was an important regional event, with implications for dissolved oxygen levels in the Gulf of Mexico, but not one that would directly impact the East Coast of the U.S.
Functions
-
logical function, public oil_tracer::register_oil_tracer(HI HI, GV GV, param_file param_file, CS CS, tr_Reg tr_Reg, restart_CS restart_CS) Register oil tracer fields and subroutines to be used with MOM.
- Parameters
[in] hi: A horizontal index type structure[in] gv: The ocean’s vertical grid structure[in] param_file: A structure to parse for run-time parameterscs: A pointer that is set to point to the control structure for this moduletr_reg: A pointer that is set to point to the control structure for the tracer advection and diffusion modulerestart_cs: A pointer to the restart control structure
-
subroutine, public oil_tracer::initialize_oil_tracer(restart restart, day day, G G, GV GV, US US, h h, diag diag, OBC OBC, CS CS, sponge_CSp sponge_CSp) Initialize the oil tracers and set up tracer output.
- Parameters
[in] restart: .true. if the fields have already been read from a restart file.[in] day: Time of the start of the run.[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[in] h: Layer thicknesses [H ~> m or kg m-2][in] diag: A structure that is used to regulate diagnostic output.obc: This open boundary condition type specifies whether, where, and what open boundary conditions are used.cs: The control structure returned by a previous call to register_oil_tracer.sponge_csp: Pointer to the control structure for the sponges.
-
subroutine, public oil_tracer::oil_tracer_column_physics(h_old h_old, h_new h_new, ea ea, eb eb, fluxes fluxes, dt dt, G G, GV GV, CS CS, tv tv, evap_CFL_limit evap_CFL_limit, minimum_forcing_depth minimum_forcing_depth) Apply sources, sinks, diapycnal mixing and rising motions to the oil tracers.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h_old: Layer thickness before entrainment [H ~> m or kg m-2].[in] h_new: Layer thickness after entrainment [H ~> m or kg m-2].[in] ea: an array to which the amount of fluid entrained[in] eb: an array to which the amount of fluid entrained[in] fluxes: A structure containing pointers to thermodynamic and tracer forcing fields. Unused fields have NULL ptrs.[in] dt: The amount of time covered by this call [s]cs: The control structure returned by a previous call to register_oil_tracer.[in] tv: A structure pointing to various thermodynamic variables[in] evap_cfl_limit: Limit on the fraction of the water that can be fluxed out of the top layer in a timestep [nondim][in] minimum_forcing_depth: The smallest depth over which fluxes can be applied [m]
-
integer function, public oil_tracer::oil_stock(h h, stocks stocks, G G, GV GV, CS CS, names names, units units, stock_index stock_index) Calculate the mass-weighted integral of the oil tracer stocks, returning the number of stocks it has calculated. If the stock_index is present, only the stock corresponding to that coded index is returned.
- Return
The number of stocks calculated here.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][out] stocks: the mass-weighted integrated amount of each tracer, in kg times concentration units [kg conc].cs: The control structure returned by a previous call to register_oil_tracer.[out] names: the names of the stocks calculated.[out] units: the units of the stocks calculated.[in] stock_index: the coded index of a specific stock being sought.
-
subroutine, public oil_tracer::oil_tracer_surface_state(state state, h h, G G, CS CS) This subroutine extracts the surface fields from this tracer package that are to be shared with the atmosphere in coupled configurations. This particular tracer package does not report anything back to the coupler.
- Parameters
[in] g: The ocean’s grid structure.[inout] state: A structure containing fields that describe the surface state of the ocean.[in] h: Layer thickness [H ~> m or kg m-2].cs: The control structure returned by a previous call to register_oil_tracer.
-
subroutine, public oil_tracer::oil_tracer_end(CS CS) Deallocate memory associated with this tracer package.
- Parameters
cs: The control structure returned by a previous call to register_oil_tracer.
Variables
-
integer, parameter oil_tracer::ntr_max = 20 the maximum number of tracers in this module.
-
-
namespace
p1m_functions¶ Linear interpolation functions.
Date of creation: 2008.06.09 L. White
This module contains p1m (linear) interpolation routines.
p1m interpolation is performed by estimating the edge values and linearly interpolating between them.
Functions
-
subroutine, public p1m_functions::p1m_interpolation(N N, h h, u u, ppoly_E ppoly_E, ppoly_coef ppoly_coef, h_neglect h_neglect) Linearly interpolate between edge values.
The resulting piecewise interpolant is stored in ‘ppoly’. See ‘ppoly.F90’ for a definition of this structure.
The edge values MUST have been estimated prior to calling this routine.
The estimated edge values must be limited to ensure monotonicity of the interpolant. We also make sure that edge values are NOT discontinuous.
It is assumed that the size of the array ‘u’ is equal to the number of cells defining ‘grid’ and ‘ppoly’. No consistency check is performed here.
- Parameters
[in] n: Number of cells[in] h: cell widths (size N)[in] u: cell average properties (size N)[inout] ppoly_e: Potentially modified edge values, with the same units as u.[inout] ppoly_coef: Potentially modified piecewise polynomial coefficients, mainly with the same units as u.[in] h_neglect: A negligibly small width in the same units as h.
-
subroutine, public p1m_functions::p1m_boundary_extrapolation(N N, h h, u u, ppoly_E ppoly_E, ppoly_coef ppoly_coef) Interpolation by linear polynomials within boundary cells.
The left and right edge values in the left and right boundary cells, respectively, are estimated using a linear extrapolation within the cells.
It is assumed that the size of the array ‘u’ is equal to the number of cells defining ‘grid’ and ‘ppoly’. No consistency check is performed here.
- Parameters
[in] n: Number of cells[in] h: cell widths (size N)[in] u: cell averages (size N)[inout] ppoly_e: edge values of piecewise polynomials, with the same units as u.[inout] ppoly_coef: coefficients of piecewise polynomials, mainly with the same units as u.
-
-
namespace
p3m_functions¶ Cubic interpolation functions.
Date of creation: 2008.06.09 L. White
This module contains p3m interpolation routines.
p3m interpolation is performed by estimating the edge values and slopes and constructing a cubic polynomial. We then make sure that the edge values are bounded and continuous and we then modify the slopes to get a monotonic cubic curve.
Functions
-
subroutine, public p3m_functions::p3m_interpolation(N N, h h, u u, ppoly_E ppoly_E, ppoly_S ppoly_S, ppoly_coef ppoly_coef, h_neglect h_neglect) Set up a piecewise cubic interpolation from cell averages and estimated edge slopes and values.
Cubic interpolation between edges.
The edge values and slopes MUST have been estimated prior to calling this routine.
It is assumed that the size of the array ‘u’ is equal to the number of cells defining ‘grid’ and ‘ppoly’. No consistency check is performed here.
- Parameters
[in] n: Number of cells[in] h: cell widths (size N)[in] u: cell averages (size N)[inout] ppoly_e: Edge value of polynomial, with the same units as u.[inout] ppoly_s: Edge slope of polynomial, in the units of u over the units of h.[inout] ppoly_coef: Coefficients of polynomial, mainly with the same units as u.[in] h_neglect: A negligibly small width for the purpose of cell reconstructions in the same units as h.
-
subroutine
p3m_limiter(N N, h h, u u, ppoly_E ppoly_E, ppoly_S ppoly_S, ppoly_coef ppoly_coef, h_neglect h_neglect)¶ Adust a piecewise cubic reconstruction with a limiter that adjusts the edge values and slopes.
The p3m limiter operates as follows:
Edge values are bounded
Discontinuous edge values are systematically averaged
Loop on cells and do the following a. Build cubic curve b. Check if cubic curve is monotonic c. If not, monotonize cubic curve and rebuild it
Step 3 of the monotonization process leaves all edge values unchanged.
- Parameters
[in] n: Number of cells[in] h: cell widths (size N)[in] u: cell averages (size N)[inout] ppoly_e: Edge value of polynomial, with the same units as u.[inout] ppoly_s: Edge slope of polynomial, in the units of u over the units of h.[inout] ppoly_coef: Coefficients of polynomial[in] h_neglect: A negligibly small width for the purpose of cell reconstructions in the same units as h.
-
subroutine, public p3m_functions::p3m_boundary_extrapolation(N N, h h, u u, ppoly_E ppoly_E, ppoly_S ppoly_S, ppoly_coef ppoly_coef, h_neglect h_neglect, h_neglect_edge h_neglect_edge) Calculate the edge values and slopes at boundary cells as part of building a piecewise cubic sub-grid scale profiles.
The following explanations apply to the left boundary cell. The same reasoning holds for the right boundary cell.
A cubic needs to be built in the cell and requires four degrees of freedom, which are the edge values and slopes. The right edge values and slopes are taken to be that of the neighboring cell (i.e., the left edge value and slope of the neighboring cell). The left edge value and slope are determined by computing the parabola based on the cell average and the right edge value and slope. The resulting cubic is not necessarily monotonic and the slopes are subsequently modified to yield a monotonic cubic.
- Parameters
[in] n: Number of cells[in] h: cell widths (size N)[in] u: cell averages (size N)[inout] ppoly_e: Edge value of polynomial, with the same units as u.[inout] ppoly_s: Edge slope of polynomial, in the units of u over the units of h.[inout] ppoly_coef: Coefficients of polynomial, mainly with the same units as u.[in] h_neglect: A negligibly small width for the purpose of cell reconstructions in the same units as h.[in] h_neglect_edge: A negligibly small width for the purpose of finding edge values in the same units as h.
-
subroutine
build_cubic_interpolant(h h, k k, ppoly_E ppoly_E, ppoly_S ppoly_S, ppoly_coef ppoly_coef)¶ Build cubic interpolant in cell k.
Given edge values and edge slopes, compute coefficients of cubic in cell k.
NOTE: edge values and slopes MUST have been properly calculated prior to calling this routine.
- Parameters
[in] h: cell widths (size N)[in] k: The index of the cell to work on[in] ppoly_e: Edge value of polynomial, with the same units as u.[in] ppoly_s: Edge slope of polynomial, in the units of u over the units of h.[inout] ppoly_coef: Coefficients of polynomial, mainly with the same units as u.
-
integer function p3m_functions::is_cubic_monotonic(ppoly_coef ppoly_coef, k k) Check whether the cubic reconstruction in cell k is monotonic.
This function checks whether the cubic curve in cell k is monotonic. If so, returns 1. Otherwise, returns 0.
The cubic is monotonic if the first derivative is single-signed in [0,1]. Hence, we check whether the roots (if any) lie inside this interval. If there is no root or if both roots lie outside this interval, the cubic is monotonic.
- Parameters
[in] ppoly_coef: Coefficients of cubic polynomial[in] k: The index of the cell to work on
-
subroutine
monotonize_cubic(h h, u0_l u0_l, u0_r u0_r, sigma_l sigma_l, sigma_r sigma_r, slope slope, u1_l u1_l, u1_r u1_r)¶ Monotonize a cubic curve by modifying the edge slopes.
This routine takes care of monotonizing a cubic on [0,1] by modifying the edge slopes. The edge values are NOT modified. The cubic is entirely determined by the four degrees of freedom u0_l, u0_r, u1_l and u1_r.
u1_l and u1_r are the edge slopes expressed in the GLOBAL coordinate system.
The monotonization occurs as follows.
- Parameters
[in] h: cell width[in] u0_l: left edge value[in] u0_r: right edge value[in] sigma_l: left 2nd-order slopes[in] sigma_r: right 2nd-order slopes[in] slope: limited PLM slope[inout] u1_l: left edge slopes[inout] u1_r: right edge slopes
Variables
-
real, parameter p3m_functions::hneglect_dflt = 1.E-30 Default value of a negligible cell thickness.
-
real, parameter p3m_functions::hneglect_edge_dflt = 1.E-10 Default value of a negligible edge thickness.
-
-
namespace
PCM_functions¶ Date of creation: 2008.06.06 L. White
This module contains routines that handle one-dimensionnal finite volume reconstruction using the piecewise constant method (PCM).
-
namespace
pcm_functions¶ Piecewise constant reconstruction functions.
Functions
-
subroutine, public pcm_functions::pcm_reconstruction(N N, u u, ppoly_E ppoly_E, ppoly_coef ppoly_coef) Reconstruction by constant polynomials within each cell. There is nothing to do but this routine is provided to ensure a homogeneous interface throughout the regridding toolbox.
It is assumed that the dimension of ‘u’ is equal to the number of cells defining ‘grid’ and ‘ppoly’. No consistency check is performed.
- Parameters
[in] n: Number of cells[in] u: cell averages[inout] ppoly_e: Edge value of polynomial, with the same units as u.[inout] ppoly_coef: Coefficients of polynomial, with the same units as u.
-
-
namespace
phillips_initialization¶ Initialization for the “Phillips” channel configuration.
By Robert Hallberg, April 1994 - June 2002
This subroutine initializes the fields for the simulations. The one argument passed to initialize, Time, is set to the current time of the simulation. The fields which are initialized here are: u - Zonal velocity [m s-1]. v - Meridional velocity [m s-1]. h - Layer thickness in m. (Must be positive.) D - Basin depth in m. (Must be positive.) f - The Coriolis parameter [s-1]. g - The reduced gravity at each interface [m s-2] Rlay - Layer potential density (coordinate variable) [kg m-3]. If ENABLE_THERMODYNAMICS is defined: T - Temperature [degC]. S - Salinity [ppt]. If SPONGE is defined: A series of subroutine calls are made to set up the damping rates and reference profiles for all variables that are damped in the sponge. Any user provided tracer code is also first linked through this subroutine.
Forcing-related fields (taux, tauy, buoy, ustar, etc.) are set in MOM_surface_forcing.F90.
These variables are all set in the set of subroutines (in this file) Phillips_initialize_thickness, Phillips_initialize_velocity, Phillips_initialize_topography and Phillips_initialize_sponges that seet up fields that are specific to the Phillips instability test case.
Functions
-
subroutine, public phillips_initialization::phillips_initialize_thickness(h h, G G, GV GV, US US, param_file param_file, just_read_params just_read_params) Initialize the thickness field for the Phillips model test case.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[out] h: The thickness that is being initialized [H ~> m or kg m-2][in] param_file: A structure indicating the open file to parse for model parameter values.[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine, public phillips_initialization::phillips_initialize_velocity(u u, v v, G G, GV GV, US US, param_file param_file, just_read_params just_read_params) Initialize the velocity fields for the Phillips model test case.
- Parameters
[in] g: Grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[out] u: i-component of velocity [m s-1][out] v: j-component of velocity [m s-1][in] param_file: A structure indicating the open file to parse for modelparameter values.[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine, public phillips_initialization::phillips_initialize_sponges(G G, GV GV, US US, tv tv, param_file param_file, CSp CSp, h h) Sets up the the inverse restoration time (Idamp), and the values towards which the interface heights and an arbitrary number of tracers should be restored within each sponge for the Phillips model test case.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[in] tv: A structure containing pointers to any available thermodynamic fields, potential temperature and salinity or mixed layer density. Absent fields have NULL ptrs.[in] param_file: A structure indicating the open file to parse for model parameter values.csp: A pointer that is set to point to the control structure for the sponge module.[in] h: Thickness field [H ~> m or kg m-2].
-
real function phillips_initialization::sech(x x) sech calculates the hyperbolic secant.
- Return
Result.
- Parameters
[in] x: Input value.
-
subroutine, public phillips_initialization::phillips_initialize_topography(D D, G G, param_file param_file, max_depth max_depth, US US) Initialize topography.
- Parameters
[in] g: The dynamic horizontal grid type[out] d: Ocean bottom depth in m or Z if US is present[in] param_file: Parameter file structure[in] max_depth: Maximum model depth in the units of D[in] us: A dimensional unit scaling type
-
-
namespace
plm_functions¶ Piecewise linear reconstruction functions.
Date of creation: 2008.06.06 L. White
This module contains routines that handle one-dimensionnal finite volume reconstruction using the piecewise linear method (PLM).
Functions
-
subroutine, public plm_functions::plm_reconstruction(N N, h h, u u, ppoly_E ppoly_E, ppoly_coef ppoly_coef, h_neglect h_neglect) Reconstruction by linear polynomials within each cell.
It is assumed that the size of the array ‘u’ is equal to the number of cells defining ‘grid’ and ‘ppoly’. No consistency check is performed here.
- Parameters
[in] n: Number of cells[in] h: cell widths (size N)[in] u: cell averages (size N)[inout] ppoly_e: edge values of piecewise polynomials, with the same units as u.[inout] ppoly_coef: coefficients of piecewise polynomials, mainly with the same units as u.[in] h_neglect: A negligibly small width for the purpose of cell reconstructions in the same units as h
-
subroutine, public plm_functions::plm_boundary_extrapolation(N N, h h, u u, ppoly_E ppoly_E, ppoly_coef ppoly_coef, h_neglect h_neglect) Reconstruction by linear polynomials within boundary cells.
The left and right edge values in the left and right boundary cells, respectively, are estimated using a linear extrapolation within the cells.
This extrapolation is EXACT when the underlying profile is linear.
It is assumed that the size of the array ‘u’ is equal to the number of cells defining ‘grid’ and ‘ppoly’. No consistency check is performed here.
- Parameters
[in] n: Number of cells[in] h: cell widths (size N)[in] u: cell averages (size N)[inout] ppoly_e: edge values of piecewise polynomials, with the same units as u.[inout] ppoly_coef: coefficients of piecewise polynomials, mainly with the same units as u.[in] h_neglect: A negligibly small width for the purpose of cell reconstructions in the same units as h
Variables
-
real, parameter plm_functions::hneglect_dflt = 1.E-30 Default negligible cell thickness.
-
-
namespace
polynomial_functions¶ Polynomial functions.
Date of creation: 2008.06.12 L. White
This module contains routines that handle polynomials.
Functions
-
real function, public polynomial_functions::evaluation_polynomial(coeff coeff, ncoef ncoef, x x) Pointwise evaluation of a polynomial at x.
The polynomial is defined by the coefficients contained in the array of the same name, as follows: C(1) + C(2)x + C(3)x^2 + C(4)x^3 + … where C refers to the array ‘coeff’. The number of coefficients is given by ncoef and x is the coordinate where the polynomial is to be evaluated.
- Parameters
[in] coeff: The coefficients of the polynomial[in] ncoef: The number of polynomial coefficients[in] x: The position at which to evaluate the polynomial
-
real function, public polynomial_functions::first_derivative_polynomial(coeff coeff, ncoef ncoef, x x) Calculates the first derivative of a polynomial evaluated at a point x.
The polynomial is defined by the coefficients contained in the array of the same name, as follows: C(1) + C(2)x + C(3)x^2 + C(4)x^3 + … where C refers to the array ‘coeff’. The number of coefficients is given by ncoef and x is the coordinate where the polynomial’s derivative is to be evaluated.
- Parameters
[in] coeff: The coefficients of the polynomial[in] ncoef: The number of polynomial coefficients[in] x: The position at which to evaluate the derivative
-
real function, public polynomial_functions::integration_polynomial(xi0 xi0, xi1 xi1, Coeff Coeff, npoly npoly) Exact integration of polynomial of degree npoly.
The array of coefficients (Coeff) must be of size npoly+1.
- Parameters
[in] xi0: The lower bound of the integral[in] xi1: The lower bound of the integral[in] coeff: The coefficients of the polynomial[in] npoly: The degree of the polynomial
-
-
namespace
ppm_functions¶ Provides functions used with the Piecewise-Parabolic-Method in the vertical ALE algorithm.
Functions
-
subroutine, public ppm_functions::ppm_reconstruction(N N, h h, u u, ppoly_E ppoly_E, ppoly_coef ppoly_coef, h_neglect h_neglect) Builds quadratic polynomials coefficients from cell mean and edge values.
- Parameters
[in] n: Number of cells[in] h: Cell widths[in] u: Cell averages[inout] ppoly_e: Edge values, with the same units as u.[inout] ppoly_coef: Polynomial coefficients, mainly with the same units as u.[in] h_neglect: A negligibly small width in the same units as h.
-
subroutine
ppm_limiter_standard(N N, h h, u u, ppoly_E ppoly_E, h_neglect h_neglect)¶ Adjusts edge values using the standard PPM limiter (Colella & Woodward, JCP 1984) after first checking that the edge values are bounded by neighbors cell averages and that the edge values are monotonic between cell averages.
- Parameters
[in] n: Number of cells[in] h: cell widths (size N)[in] u: cell average properties (size N)[inout] ppoly_e: Potentially modified edge values, with the same units as u.[in] h_neglect: A negligibly small width in the same units as h.
-
subroutine, public ppm_functions::ppm_boundary_extrapolation(N N, h h, u u, ppoly_E ppoly_E, ppoly_coef ppoly_coef, h_neglect h_neglect) Reconstruction by parabolas within boundary cells.
- Parameters
[in] n: Number of cells[in] h: cell widths (size N)[in] u: cell averages (size N)[inout] ppoly_e: edge values of piecewise polynomials, with the same units as u.[inout] ppoly_coef: coefficients of piecewise polynomials, mainly with the same units as u.[in] h_neglect: A negligibly small width for the purpose of cell reconstructions in the same units as h.
Variables
-
real, parameter ppm_functions::hneglect_dflt = 1.E-30 A tiny width that is so small that adding it to cell widths does not change the value due to a computational representation. It is used to avoid division by zero.
- Note
This is a dimensional parameter and should really include a unit conversion.
-
-
namespace
pqm_functions¶ Piecewise quartic reconstruction functions.
Date of creation: 2008.06.06 L. White
This module contains routines that handle one-dimensionnal finite volume reconstruction using the piecewise quartic method (PQM).
Functions
-
subroutine, public pqm_functions::pqm_reconstruction(N N, h h, u u, ppoly_E ppoly_E, ppoly_S ppoly_S, ppoly_coef ppoly_coef, h_neglect h_neglect) Reconstruction by quartic polynomials within each cell.
It is assumed that the dimension of ‘u’ is equal to the number of cells defining ‘grid’ and ‘ppoly’. No consistency check is performed.
- Parameters
[in] n: Number of cells[in] h: cell widths (size N)[in] u: cell averages (size N)[inout] ppoly_e: Edge value of polynomial, with the same units as u.[inout] ppoly_s: Edge slope of polynomial, in the units of u over the units of h.[inout] ppoly_coef: Coefficients of polynomial, mainly with the same units as u.[in] h_neglect: A negligibly small width for the purpose of cell reconstructions in the same units as h
-
subroutine
pqm_limiter(N N, h h, u u, ppoly_E ppoly_E, ppoly_S ppoly_S, h_neglect h_neglect)¶ Limit the piecewise quartic method reconstruction.
Standard PQM limiter (White & Adcroft, JCP 2008).
It is assumed that the dimension of ‘u’ is equal to the number of cells defining ‘grid’ and ‘ppoly’. No consistency check is performed.
- Parameters
[in] n: Number of cells[in] h: cell widths (size N)[in] u: cell average properties (size N)[inout] ppoly_e: Potentially modified edge values, with the same units as u.[inout] ppoly_s: Potentially modified edge slopes, with the same units as u.[in] h_neglect: A negligibly small width for the purpose of cell reconstructions in the same units as h
-
subroutine, public pqm_functions::pqm_boundary_extrapolation(N N, h h, u u, ppoly_E ppoly_E, ppoly_coef ppoly_coef) Reconstruction by parabolas within boundary cells.
The following explanations apply to the left boundary cell. The same reasoning holds for the right boundary cell.
A parabola needs to be built in the cell and requires three degrees of freedom, which are the right edge value and slope and the cell average. The right edge values and slopes are taken to be that of the neighboring cell (i.e., the left edge value and slope of the neighboring cell). The resulting parabola is not necessarily monotonic and the traditional PPM limiter is used to modify one of the edge values in order to yield a monotonic parabola.
It is assumed that the size of the array ‘u’ is equal to the number of cells defining ‘grid’ and ‘ppoly’. No consistency check is performed here.
- Parameters
[in] n: Number of cells[in] h: cell widths (size N)[in] u: cell averages (size N)[inout] ppoly_e: Edge value of polynomial, with the same units as u.[inout] ppoly_coef: Coefficients of polynomial, mainly with the same units as u.
-
subroutine, public pqm_functions::pqm_boundary_extrapolation_v1(N N, h h, u u, ppoly_E ppoly_E, ppoly_S ppoly_S, ppoly_coef ppoly_coef, h_neglect h_neglect) Reconstruction by parabolas within boundary cells.
The following explanations apply to the left boundary cell. The same reasoning holds for the right boundary cell.
A parabola needs to be built in the cell and requires three degrees of freedom, which are the right edge value and slope and the cell average. The right edge values and slopes are taken to be that of the neighboring cell (i.e., the left edge value and slope of the neighboring cell). The resulting parabola is not necessarily monotonic and the traditional PPM limiter is used to modify one of the edge values in order to yield a monotonic parabola.
It is assumed that the size of the array ‘u’ is equal to the number of cells defining ‘grid’ and ‘ppoly’. No consistency check is performed here.
- Parameters
[in] n: Number of cells[in] h: cell widths (size N)[in] u: cell averages (size N)[inout] ppoly_e: Edge value of polynomial, with the same units as u.[inout] ppoly_s: Edge slope of polynomial, in the units of u over the units of h.[inout] ppoly_coef: Coefficients of polynomial, mainly with the same units as u.[in] h_neglect: A negligibly small width for the purpose of cell reconstructions in the same units as h.
Variables
-
real, parameter pqm_functions::hneglect_dflt = 1.E-30 Default negligible cell thickness.
-
-
namespace
pseudo_salt_tracer¶ A tracer package that mimics salinity.
By Andrew Shao, 2016
This file contains the routines necessary to model a passive tracer that uses the same boundary fluxes as salinity. At the beginning of the run, salt is set to the same as tvS. Any deviations between this salt-like tracer and tvS signifies a difference between how active and passive tracers are treated.
Functions
-
logical function, public pseudo_salt_tracer::register_pseudo_salt_tracer(HI HI, GV GV, param_file param_file, CS CS, tr_Reg tr_Reg, restart_CS restart_CS) Register the pseudo-salt tracer with MOM6.
- Parameters
[in] hi: A horizontal index type structure[in] gv: The ocean’s vertical grid structure[in] param_file: A structure to parse for run-time parameterscs: The control structure returned by a previous call to register_pseudo_salt_tracer.tr_reg: A pointer that is set to point to the control structure for the tracer advection and diffusion modulerestart_cs: A pointer to the restart control structure
-
subroutine, public pseudo_salt_tracer::initialize_pseudo_salt_tracer(restart restart, day day, G G, GV GV, h h, diag diag, OBC OBC, CS CS, sponge_CSp sponge_CSp, tv tv) Initialize the pseudo-salt tracer.
- Parameters
[in] restart: .true. if the fields have already been read from a restart file.[in] day: Time of the start of the run.[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][in] diag: A structure that is used to regulate diagnostic output.obc: This open boundary condition type specifies whether, where, and what open boundary conditions are used.cs: The control structure returned by a previous call to register_pseudo_salt_tracer.sponge_csp: Pointer to the control structure for the sponges.[in] tv: A structure pointing to various thermodynamic variables
-
subroutine, public pseudo_salt_tracer::pseudo_salt_tracer_column_physics(h_old h_old, h_new h_new, ea ea, eb eb, fluxes fluxes, dt dt, G G, GV GV, CS CS, tv tv, debug debug, evap_CFL_limit evap_CFL_limit, minimum_forcing_depth minimum_forcing_depth) Apply sources, sinks and diapycnal diffusion to the tracers in this package.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h_old: Layer thickness before entrainment [H ~> m or kg m-2].[in] h_new: Layer thickness after entrainment [H ~> m or kg m-2].[in] ea: an array to which the amount of fluid entrained[in] eb: an array to which the amount of fluid entrained[in] fluxes: A structure containing pointers to thermodynamic and tracer forcing fields. Unused fields have NULL ptrs.[in] dt: The amount of time covered by this call [s]cs: The control structure returned by a previous call to register_pseudo_salt_tracer.[in] tv: A structure pointing to various thermodynamic variables[in] debug: If true calculate checksums[in] evap_cfl_limit: Limit on the fraction of the water that can be fluxed out of the top layer in a timestep [nondim][in] minimum_forcing_depth: The smallest depth over which fluxes can be applied [m]
-
integer function, public pseudo_salt_tracer::pseudo_salt_stock(h h, stocks stocks, G G, GV GV, CS CS, names names, units units, stock_index stock_index) Calculates the mass-weighted integral of all tracer stocks, returning the number of stocks it has calculated. If the stock_index is present, only the stock corresponding to that coded index is returned.
- Return
Return value: the number of stocks calculated here.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][out] stocks: the mass-weighted integrated amount of each tracer, in kg times concentration units [kg conc].cs: The control structure returned by a previous call to register_pseudo_salt_tracer.[out] names: The names of the stocks calculated.[out] units: The units of the stocks calculated.[in] stock_index: The coded index of a specific stock being sought.
-
subroutine, public pseudo_salt_tracer::pseudo_salt_tracer_surface_state(state state, h h, G G, CS CS) This subroutine extracts the surface fields from this tracer package that are to be shared with the atmosphere in coupled configurations. This particular tracer package does not report anything back to the coupler.
- Parameters
[in] g: The ocean’s grid structure.[inout] state: A structure containing fields that describe the surface state of the ocean.[in] h: Layer thickness [H ~> m or kg m-2].cs: The control structure returned by a previous call to register_pseudo_salt_tracer.
-
subroutine, public pseudo_salt_tracer::pseudo_salt_tracer_end(CS CS) Deallocate memory associated with this tracer package.
- Parameters
cs: The control structure returned by a previous call to register_pseudo_salt_tracer.
-
-
namespace
regional_dyes¶ A tracer package for using dyes to diagnose regional flows.
This file contains an example of the code that is needed to set up and use a set (in this case two) of dynamically passive tracers for diagnostic purposes. The tracers here are dye tracers which are set to 1 within the geographical region specified. The depth which a tracer is set is determined by calculating the depth from the seafloor upwards through the column.
Functions
-
logical function, public regional_dyes::register_dye_tracer(HI HI, GV GV, US US, param_file param_file, CS CS, tr_Reg tr_Reg, restart_CS restart_CS) This subroutine is used to register tracer fields and subroutines to be used with MOM.
- Parameters
[in] hi: A horizontal index type structure.[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameterscs: A pointer that is set to point to the control structure for this moduletr_reg: A pointer that is set to point to the control structure for the tracer advection and diffusion module.restart_cs: A pointer to the restart control structure.
-
subroutine, public regional_dyes::initialize_dye_tracer(restart restart, day day, G G, GV GV, h h, diag diag, OBC OBC, CS CS, sponge_CSp sponge_CSp) This subroutine initializes the CSntr tracer fields in tr(:,:,:,:) and it sets up the tracer output.
- Parameters
[in] restart: .true. if the fields have already been read from a restart file.[in] day: Time of the start of the run.[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][in] diag: Structure used to regulate diagnostic output.obc: This open boundary condition type specifies whether, where, and what open boundary conditions are used.cs: The control structure returned by a previous call to register_dye_tracer.sponge_csp: A pointer to the control structure for the sponges, if they are in use.
-
subroutine, public regional_dyes::dye_tracer_column_physics(h_old h_old, h_new h_new, ea ea, eb eb, fluxes fluxes, dt dt, G G, GV GV, CS CS, evap_CFL_limit evap_CFL_limit, minimum_forcing_depth minimum_forcing_depth) This subroutine applies diapycnal diffusion and any other column tracer physics or chemistry to the tracers from this file. This is a simple example of a set of advected passive tracers. The arguments to this subroutine are redundant in that h_new(k) = h_old(k) + ea(k) - eb(k-1) + eb(k) - ea(k+1)
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h_old: Layer thickness before entrainment [H ~> m or kg m-2].[in] h_new: Layer thickness after entrainment [H ~> m or kg m-2].[in] ea: an array to which the amount of fluid entrained[in] eb: an array to which the amount of fluid entrained[in] fluxes: A structure containing pointers to thermodynamic and tracer forcing fields. Unused fields have NULL ptrs.[in] dt: The amount of time covered by this call [s]cs: The control structure returned by a previous call to register_dye_tracer.[in] evap_cfl_limit: Limit on the fraction of the water that can be fluxed out of the top layer in a timestep [nondim][in] minimum_forcing_depth: The smallest depth over which fluxes can be applied [m]
-
integer function, public regional_dyes::dye_stock(h h, stocks stocks, G G, GV GV, CS CS, names names, units units, stock_index stock_index) This function calculates the mass-weighted integral of all tracer stocks, returning the number of stocks it has calculated. If the stock_index is present, only the stock corresponding to that coded index is returned.
- Return
Return value: the number of stocks calculated here.
- Parameters
[in] h: Layer thicknesses [H ~> m or kg m-2][out] stocks: the mass-weighted integrated amount of each tracer, in kg times concentration units [kg conc].[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structurecs: The control structure returned by a previous call to register_dye_tracer.[out] names: the names of the stocks calculated.[out] units: the units of the stocks calculated.[in] stock_index: the coded index of a specific stock being sought.
-
subroutine, public regional_dyes::dye_tracer_surface_state(state state, h h, G G, CS CS) This subroutine extracts the surface fields from this tracer package that are to be shared with the atmosphere in coupled configurations. This particular tracer package does not report anything back to the coupler.
- Parameters
[in] g: The ocean’s grid structure.[inout] state: A structure containing fields that describe the surface state of the ocean.[in] h: Layer thickness [H ~> m or kg m-2].cs: The control structure returned by a previous call to register_dye_tracer.
-
subroutine, public regional_dyes::regional_dyes_end(CS CS) Clean up any allocated memory after the run.
- Parameters
cs: The control structure returned by a previous call to register_dye_tracer.
-
-
namespace
regrid_consts¶ Contains constants for interpreting input parameters that control regridding.
Functions
-
integer function regrid_consts::coordinatemode(string string) Parse a string parameter specifying the coordinate mode and return the appropriate enumerated integer.
- Return
Enumerated integer indicating coordinate mode
- Parameters
[in] string: String to indicate coordinate mode
-
character(len=16) function regrid_consts::coordinateunitsi(coordMode coordMode) Returns a string with the coordinate units associated with the enumerated integer,.
- Return
Units of coordinate
- Parameters
[in] coordmode: Coordinate mode
-
character(len=16) function regrid_consts::coordinateunitss(string string) Returns a string with the coordinate units associated with the string defining the coordinate mode.
- Return
Units of coordinate
- Parameters
[in] string: Coordinate mode
-
logical function regrid_consts::state_dependent_char(string string) Returns true if the coordinate is dependent on the state density, returns false otherwise.
- Parameters
[in] string: String to indicate coordinate mode
-
logical function regrid_consts::state_dependent_int(mode mode) Returns true if the coordinate is dependent on the state density, returns false otherwise.
- Parameters
[in] mode: Coordinate mode
Variables
-
integer, parameter regrid_consts::regridding_layer = 1 Layer mode identifier.
-
integer, parameter regrid_consts::regridding_zstar = 2 z* coordinates identifier
-
integer, parameter regrid_consts::regridding_rho = 3 Density coordinates identifier.
-
integer, parameter regrid_consts::regridding_sigma = 4 Sigma coordinates identifier.
-
integer, parameter regrid_consts::regridding_arbitrary = 5 Arbitrary coordinates identifier.
-
integer, parameter regrid_consts::regridding_hycom1 = 6 Simple HyCOM coordinates without BBL.
-
integer, parameter regrid_consts::regridding_slight = 7 Identifier for stretched coordinates in the lightest water, isopycnal below.
-
integer, parameter regrid_consts::regridding_sigma_shelf_zstar = 8 Identifiered for z* coordinates at the bottom, sigma-near the top.
-
integer, parameter regrid_consts::regridding_adaptive = 9 Adaptive coordinate mode identifier.
-
character(len=*), parameter regrid_consts::regridding_layer_string = "LAYER" Layer string.
-
character(len=*), parameter regrid_consts::regridding_zstar_string_old = "Z*" z* string (legacy name)
-
character(len=*), parameter regrid_consts::regridding_zstar_string = "ZSTAR" z* string
-
character(len=*), parameter regrid_consts::regridding_rho_string = "RHO" Rho string.
-
character(len=*), parameter regrid_consts::regridding_sigma_string = "SIGMA" Sigma string.
-
character(len=*), parameter regrid_consts::regridding_arbitrary_string = "ARB" Arbitrary coordinates.
-
character(len=*), parameter regrid_consts::regridding_hycom1_string = "HYCOM1" Hycom string.
-
character(len=*), parameter regrid_consts::regridding_slight_string = "SLIGHT" Hybrid S-rho string.
-
character(len=*), parameter regrid_consts::regridding_sigma_shelf_zstar_string = "SIGMA_SHELF_ZSTAR" Hybrid z*/sigma.
-
character(len=*), parameter regrid_consts::regridding_adaptive_string = "ADAPTIVE" Adaptive coordinate string.
-
character(len=*), parameter regrid_consts::default_coordinate_mode = REGRIDDING_LAYER_STRING Default coordinate mode.
-
-
namespace
regrid_edge_slopes¶ Routines that estimate edge slopes to be used in high-order reconstruction schemes.
Functions
-
subroutine, public regrid_edge_slopes::edge_slopes_implicit_h3(N N, h h, u u, edge_slopes edge_slopes, h_neglect h_neglect) Compute ih4 edge slopes (implicit third order accurate) in the same units as h.
Compute edge slopes based on third-order implicit estimates. Note that the estimates are fourth-order accurate on uniform grids
Third-order implicit estimates of edge slopes are based on a two-cell stencil. A tridiagonal system is set up and is based on expressing the edge slopes in terms of neighboring cell averages. The generic relationship is
\[ \alpha u'_{i-1/2} + u'_{i+1/2} + \beta u'_{i+3/2} = a \bar{u}_i + b \bar{u}_{i+1} \]and the stencil looks like this
..o—o—o.. i-1/2 i+1/2 i+3/2i i+1
In this routine, the coefficients \(\alpha\), \(\beta\), a and b are computed, the tridiagonal system is built, boundary conditions are prescribed and the system is solved to yield edge-slope estimates.
There are N+1 unknowns and we are able to write N-1 equations. The boundary conditions close the system.
- Parameters
[in] n: Number of cells[in] h: cell widths (size N)[in] u: cell average properties (size N)[inout] edge_slopes: Returned edge slopes, with the same units as u divided by the units of h.[in] h_neglect: A negligibly small width
-
subroutine, public regrid_edge_slopes::edge_slopes_implicit_h5(N N, h h, u u, edge_slopes edge_slopes, h_neglect h_neglect) Compute ih5 edge values (implicit fifth order accurate)
- Parameters
[in] n: Number of cells[in] h: cell widths (size N)[in] u: cell average properties (size N)[inout] edge_slopes: Returned edge slopes, with the same units as u divided by the units of h.[in] h_neglect: A negligibly small width in the same units as h.
Variables
-
real, parameter regrid_edge_slopes::hneglect_dflt = 1.E-30 Default negligible cell thickness.
-
-
namespace
regrid_edge_values¶ Edge value estimation for high-order resconstruction.
Functions
-
subroutine, public regrid_edge_values::bound_edge_values(N N, h h, u u, edge_val edge_val, h_neglect h_neglect) Bound edge values by neighboring cell averages.
In this routine, we loop on all cells to bound their left and right edge values by the cell averages. That is, the left edge value must lie between the left cell average and the central cell average. A similar reasoning applies to the right edge values.
Both boundary edge values are set equal to the boundary cell averages. Any extrapolation scheme is applied after this routine has been called. Therefore, boundary cells are treated as if they were local extrama.
- Parameters
[in] n: Number of cells[in] h: cell widths (size N)[in] u: cell average properties (size N)[inout] edge_val: Potentially modified edge values, with the same units as u.[in] h_neglect: A negligibly small width in the same units as h.
-
subroutine, public regrid_edge_values::average_discontinuous_edge_values(N N, edge_val edge_val) Replace discontinuous collocated edge values with their average.
For each interior edge, check whether the edge values are discontinuous. If so, compute the average and replace the edge values by the average.
- Parameters
[in] n: Number of cells[inout] edge_val: Edge values that may be modified the second index size is 2.
-
subroutine, public regrid_edge_values::check_discontinuous_edge_values(N N, u u, edge_val edge_val) Check discontinuous edge values and replace them with their average if not monotonic.
For each interior edge, check whether the edge values are discontinuous. If so and if they are not monotonic, replace each edge value by their average.
- Parameters
[in] n: Number of cells[in] u: cell averages (size N)[inout] edge_val: Cell edge values with the same units as u.
-
subroutine, public regrid_edge_values::edge_values_explicit_h2(N N, h h, u u, edge_val edge_val, h_neglect h_neglect) Compute h2 edge values (explicit second order accurate) in the same units as h.
- Parameters
[in] n: Number of cells[in] h: cell widths (size N)[in] u: cell average properties (size N)[inout] edge_val: Returned edge values, with the same units as u; the second index size is 2.[in] h_neglect: A negligibly small width
-
subroutine, public regrid_edge_values::edge_values_explicit_h4(N N, h h, u u, edge_val edge_val, h_neglect h_neglect) Compute h4 edge values (explicit fourth order accurate) in the same units as h.
Compute edge values based on fourth-order explicit estimates. These estimates are based on a cubic interpolant spanning four cells and evaluated at the location of the middle edge. An interpolant spanning cells i-2, i-1, i and i+1 is evaluated at edge i-1/2. The estimate for each edge is unique.
..o—o—o—o—o.. i-1/2i-2 i-1 i i+1
The first two edge values are estimated by evaluating the first available cubic interpolant, i.e., the interpolant spanning cells 1, 2, 3 and 4. Similarly, the last two edge values are estimated by evaluating the last available interpolant.
For this fourth-order scheme, at least four cells must exist.
- Parameters
[in] n: Number of cells[in] h: cell widths (size N)[in] u: cell average properties (size N)[inout] edge_val: Returned edge values, with the same units as u; the second index size is 2.[in] h_neglect: A negligibly small width
-
subroutine, public regrid_edge_values::edge_values_implicit_h4(N N, h h, u u, edge_val edge_val, h_neglect h_neglect) Compute ih4 edge values (implicit fourth order accurate) in the same units as h.
Compute edge values based on fourth-order implicit estimates.
Fourth-order implicit estimates of edge values are based on a two-cell stencil. A tridiagonal system is set up and is based on expressing the edge values in terms of neighboring cell averages. The generic relationship is
\[ \alpha u_{i-1/2} + u_{i+1/2} + \beta u_{i+3/2} = a \bar{u}_i + b \bar{u}_{i+1} \]and the stencil looks like this
..o—o—o.. i-1/2 i+1/2 i+3/2i i+1
In this routine, the coefficients \(\alpha\), \(\beta\), \(a\) and \(b\) are computed, the tridiagonal system is built, boundary conditions are prescribed and the system is solved to yield edge-value estimates.
There are N+1 unknowns and we are able to write N-1 equations. The boundary conditions close the system.
- Parameters
[in] n: Number of cells[in] h: cell widths (size N)[in] u: cell average properties (size N)[inout] edge_val: Returned edge values, with the same units as u; the second index size is 2.[in] h_neglect: A negligibly small width
-
subroutine, public regrid_edge_values::edge_values_implicit_h6(N N, h h, u u, edge_val edge_val, h_neglect h_neglect) Compute ih6 edge values (implicit sixth order accurate) in the same units as h.
Sixth-order implicit estimates of edge values are based on a four-cell, three-edge stencil. A tridiagonal system is set up and is based on expressing the edge values in terms of neighboring cell averages.
The generic relationship is
\[ \alpha u_{i-1/2} + u_{i+1/2} + \beta u_{i+3/2} = a \bar{u}_{i-1} + b \bar{u}_i + c \bar{u}_{i+1} + d \bar{u}_{i+2} \]and the stencil looks like this
..o—o—o—o—o.. i-1/2 i+1/2 i+3/2i-1 i i+1 i+2
In this routine, the coefficients \(\alpha\), \(\beta\), a, b, c and d are computed, the tridiagonal system is built, boundary conditions are prescribed and the system is solved to yield edge-value estimates.
Note that the centered stencil only applies to edges 3 to N-1 (edges are numbered 1 to n+1), which yields N-3 equations for N+1 unknowns. Two other equations are written by using a right-biased stencil for edge 2 and a left-biased stencil for edge N. The prescription of boundary conditions (using sixth-order polynomials) closes the system.
CAUTION: For each edge, in order to determine the coefficients of the implicit expression, a 6x6 linear system is solved. This may become computationally expensive if regridding is carried out often. Figuring out closed-form expressions for these coefficients on nonuniform meshes turned out to be intractable.
- Parameters
[in] n: Number of cells[in] h: cell widths (size N)[in] u: cell average properties (size N)[inout] edge_val: Returned edge values, with the same units as u; the second index size is 2.[in] h_neglect: A negligibly small width
Variables
-
real, parameter regrid_edge_values::hneglect_edge_dflt = 1.e-10 The default value for cut-off minimum thickness for sum(h) in edge value inversions.
-
real, parameter regrid_edge_values::hneglect_dflt = 1.e-30 The default value for cut-off minimum thickness for sum(h) in other calculations.
-
real, parameter regrid_edge_values::hminfrac = 1.e-5 A minimum fraction for min(h)/sum(h)
-
-
namespace
regrid_interp¶ Vertical interpolation for regridding.
Unnamed Group
-
integer, parameter regrid_interp::degree_1 = 1 Interpolant degrees.
-
integer, parameter regrid_interp::degree_2 = 2 Interpolant degrees.
-
integer, parameter regrid_interp::degree_3 = 3 Interpolant degrees.
-
integer, parameter regrid_interp::degree_4 = 4 Interpolant degrees.
-
integer, parameter, public regrid_interp::degree_max = 5 Interpolant degrees.
-
real, parameter, public regrid_interp::nr_offset = 1e-6 When the N-R algorithm produces an estimate that lies outside [0,1], the estimate is set to be equal to the boundary location, 0 or 1, plus or minus an offset, respectively, when the derivative is zero at the boundary.
-
integer, parameter, public regrid_interp::nr_iterations = 8 Maximum number of Newton-Raphson iterations. Newton-Raphson iterations are used to build the new grid by finding the coordinates associated with target densities and interpolations of degree larger than 1.
-
real, parameter, public regrid_interp::nr_tolerance = 1e-12 Tolerance for Newton-Raphson iterations (stop when increment falls below this)
-
subroutine, public regrid_interp::regridding_set_ppolys(CS CS, densities densities, n0 n0, h0 h0, ppoly0_E ppoly0_E, ppoly0_S ppoly0_S, ppoly0_coefs ppoly0_coefs, degree degree, h_neglect h_neglect, h_neglect_edge h_neglect_edge) Builds an interpolated profile for the densities within each grid cell.
It may happen that, given a high-order interpolator, the number of available layers is insufficient (e.g., there are two available layers for a third-order PPM ih4 scheme). In these cases, we resort to the simplest continuous linear scheme (P1M h2).
- Parameters
[in] cs: Interpolation control structure[in] densities: Actual cell densities[in] n0: Number of cells on source grid[in] h0: cell widths on source grid[inout] ppoly0_e: Edge value of polynomial[inout] ppoly0_s: Edge slope of polynomial[inout] ppoly0_coefs: Coefficients of polynomial[inout] degree: The degree of the polynomials[in] h_neglect: A negligibly small width for the purpose of cell reconstructions in the same units as h0.[in] h_neglect_edge: A negligibly small width for the purpose of edge value calculations in the same units as h0.
-
subroutine, public regrid_interp::interpolate_grid(n0 n0, h0 h0, x0 x0, ppoly0_E ppoly0_E, ppoly0_coefs ppoly0_coefs, target_values target_values, degree degree, n1 n1, h1 h1, x1 x1) Given target values (e.g., density), build new grid based on polynomial.
Given the grid ‘grid0’ and the piecewise polynomial interpolant ‘ppoly0’ (possibly discontinuous), the coordinates of the new grid ‘grid1’ are determined by finding the corresponding target interface densities.
- Parameters
[in] n0: Number of points on source grid[in] h0: Thicknesses of source grid cells[in] x0: Source interface positions[in] ppoly0_e: Edge values of interpolating polynomials[in] ppoly0_coefs: Coefficients of interpolating polynomials[in] target_values: Target values of interfaces[in] degree: Degree of interpolating polynomials[in] n1: Number of points on target grid[inout] h1: Thicknesses of target grid cells[inout] x1: Target interface positions
-
subroutine, public regrid_interp::build_and_interpolate_grid(CS CS, densities densities, n0 n0, h0 h0, x0 x0, target_values target_values, n1 n1, h1 h1, x1 x1, h_neglect h_neglect, h_neglect_edge h_neglect_edge) Build a grid by interpolating for target values.
- Parameters
[in] cs: A control structure for regrid_interp[in] densities: Input cell densities [kg m-3][in] target_values: Target values of interfaces[in] n0: The number of points on the input grid[in] h0: Initial cell widths[in] x0: Source interface positions[in] n1: The number of points on the output grid[inout] h1: Output cell widths[inout] x1: Target interface positions[in] h_neglect: A negligibly small width for the purpose of cell reconstructions in the same units as h0.[in] h_neglect_edge: A negligibly small width for the purpose of edge value calculations in the same units as h0.
-
real function regrid_interp::get_polynomial_coordinate(N N, h h, x_g x_g, ppoly_E ppoly_E, ppoly_coefs ppoly_coefs, target_value target_value, degree degree) Given a target value, find corresponding coordinate for given polynomial.
Here, ‘ppoly’ is assumed to be a piecewise discontinuous polynomial of degree ‘degree’ throughout the domain defined by ‘grid’. A target value is given and we need to determine the corresponding grid coordinate to define the new grid.
If the target value is out of range, the grid coordinate is simply set to be equal to one of the boundary coordinates, which results in vanished layers near the boundaries.
IT IS ASSUMED THAT THE PIECEWISE POLYNOMIAL IS MONOTONICALLY INCREASING. IF THIS IS NOT THE CASE, THE NEW GRID MAY BE ILL-DEFINED.
It is assumed that the number of cells defining ‘grid’ and ‘ppoly’ are the same.
- Return
The position of x_g at which target_value is found.
- Parameters
[in] n: Number of grid cells[in] h: Grid cell thicknesses[in] x_g: Grid interface locations[in] ppoly_e: Edge values of interpolating polynomials[in] ppoly_coefs: Coefficients of interpolating polynomials[in] target_value: Target value to find position for[in] degree: Degree of the interpolating polynomials
-
integer function regrid_interp::interpolation_scheme(interp_scheme interp_scheme) Numeric value of interpolation_scheme corresponding to scheme name.
- Parameters
[in] interp_scheme: Name of the interpolation scheme Valid values include “P1M_H2”, “P1M_H4”, “P1M_IH2”, “PLM”, “PPM_H4”, “PPM_IH4”, “P3M_IH4IH3”, “P3M_IH6IH5”, “PQM_IH4IH3”, and “PQM_IH6IH5”
-
subroutine, public regrid_interp::set_interp_scheme(CS CS, interp_scheme interp_scheme) Store the interpolation_scheme value in the interp_CS based on the input string.
- Parameters
[inout] cs: A control structure for regrid_interp[in] interp_scheme: Name of the interpolation scheme Valid values include “P1M_H2”, “P1M_H4”, “P1M_IH2”, “PLM”, “PPM_H4”, “PPM_IH4”, “P3M_IH4IH3”, “P3M_IH6IH5”, “PQM_IH4IH3”, and “PQM_IH6IH5”
-
subroutine, public regrid_interp::set_interp_extrap(CS CS, extrap extrap) Store the boundary_extrapolation value in the interp_CS.
- Parameters
[inout] cs: A control structure for regrid_interp[in] extrap: Indicate whether high-order boundary extrapolation should be used in boundary cells
Variables
-
integer, parameter regrid_interp::interpolation_p1m_h2 = 0 O(h^2)
-
integer, parameter regrid_interp::interpolation_p1m_h4 = 1 O(h^2)
-
integer, parameter regrid_interp::interpolation_p1m_ih4 = 2 O(h^2)
-
integer, parameter regrid_interp::interpolation_plm = 3 O(h^2)
-
integer, parameter regrid_interp::interpolation_ppm_h4 = 4 O(h^3)
-
integer, parameter regrid_interp::interpolation_ppm_ih4 = 5 O(h^3)
-
integer, parameter regrid_interp::interpolation_p3m_ih4ih3 = 6 O(h^4)
-
integer, parameter regrid_interp::interpolation_p3m_ih6ih5 = 7 O(h^4)
-
integer, parameter regrid_interp::interpolation_pqm_ih4ih3 = 8 O(h^4)
-
integer, parameter regrid_interp::interpolation_pqm_ih6ih5 = 9 O(h^5)
-
-
namespace
regrid_solvers¶ Solvers of linear systems.
Date of creation: 2008.06.12 L. White
This module contains solvers of linear systems. These routines could (should ?) be replaced later by more efficient ones.
Functions
-
subroutine, public regrid_solvers::solve_linear_system(A A, B B, X X, system_size system_size) Solve the linear system AX = B by Gaussian elimination.
This routine uses Gauss’s algorithm to transform the system’s original matrix into an upper triangular matrix. Back substitution yields the answer. The matrix A must be square and its size must be that of the vectors B and X.
- Parameters
[inout] a: The matrix being inverted[inout] b: system right-hand side[inout] x: solution vector[in] system_size: The size of the system
-
subroutine, public regrid_solvers::solve_tridiagonal_system(Al Al, Ad Ad, Au Au, B B, X X, system_size system_size) Solve the tridiagonal system AX = B.
This routine uses Thomas’s algorithm to solve the tridiagonal system AX = B. (A is made up of lower, middle and upper diagonals)
- Parameters
[inout] ad: Maxtix center diagonal[inout] al: Matrix lower diagonal[inout] au: Matrix upper diagonal[inout] b: system right-hand side[inout] x: solution vector[in] system_size: The size of the system
-
-
namespace
rgc_initialization¶ Functions
-
subroutine, public rgc_initialization::rgc_initialize_sponges(G G, GV GV, tv tv, u u, v v, PF PF, use_ALE use_ALE, CSp CSp, ACSp ACSp) Sets up the the inverse restoration time (Idamp), and.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] tv: A structure containing pointers to any available thermodynamic fields, potential temperature and salinity or mixed layer density. Absent fields have NULL ptrs.[in] u: u velocity.[in] v: v velocity.[in] pf: A structure indicating the open file to parse for model parameter values.[in] use_ale: If true, indicates model is in ALE modecsp: Layer-mode sponge structureacsp: ALE-mode sponge structure
Variables
-
character(len=40) rgc_initialization::mod = "RGC_initialization"
-
-
namespace
rgc_tracer¶ This module contains the routines used to set up a dynamically passive tracer. Set up and use passive tracers requires the following: (1) register_RGC_tracer (2) apply diffusion, physics/chemistry and advect the tracer.
Functions
-
logical function, public rgc_tracer::register_rgc_tracer(HI HI, GV GV, param_file param_file, CS CS, tr_Reg tr_Reg, restart_CS restart_CS) This subroutine is used to register tracer fields.
- Parameters
[in] hi: A horizontal index type structure.[in] gv: The ocean’s vertical grid structure.[in] param_file: A structure indicating the open file to parse for model parameter values.cs: A pointer that is set to point to the control structure for this module (in/out).tr_reg: A pointer to the tracer registry.restart_cs: A pointer to the restart control structure.
-
subroutine, public rgc_tracer::initialize_rgc_tracer(restart restart, day day, G G, GV GV, h h, diag diag, OBC OBC, CS CS, layer_CSp layer_CSp, sponge_CSp sponge_CSp) Initializes the NTR tracer fields in tr(:,:,:,:) and it sets up the tracer output.
- Parameters
[in] g: Grid structure.[in] gv: The ocean’s vertical grid structure.[in] restart: .true. if the fields have already been read from a restart file.[in] day: Time of the start of the run.[in] h: Layer thickness, in m or kg m-2.[in] diag: Structure used to regulate diagnostic output.obc: This open boundary condition type specifies whether, where, and what open boundary conditions are used. This is not being used for now.cs: The control structure returned by a previous call to RGC_register_tracer.layer_csp: A pointer to the control structuresponge_csp: A pointer to the control structure for the sponges, if they are in use. Otherwise this may be unassociated.
-
subroutine, public rgc_tracer::rgc_tracer_column_physics(h_old h_old, h_new h_new, ea ea, eb eb, fluxes fluxes, dt dt, G G, GV GV, CS CS, evap_CFL_limit evap_CFL_limit, minimum_forcing_depth minimum_forcing_depth) This subroutine applies diapycnal diffusion and any other column tracer physics or chemistry to the tracers from this file. This is a simple example of a set of advected passive tracers.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] h_old: Layer thickness before entrainment [H ~> m or kg m-2].[in] h_new: Layer thickness after entrainment [H ~> m or kg m-2].[in] ea: an array to which the amount of fluid entrained[in] eb: an array to which the amount of fluid entrained[in] fluxes: A structure containing pointers to any possible forcing fields. Unused fields have NULL ptrs.[in] dt: The amount of time covered by this call [s].cs: The control structure returned by a previous call.[in] evap_cfl_limit: Limit on the fraction of the water that can be fluxed out of the top layer in a timestep [nondim].[in] minimum_forcing_depth: The smallest depth over which fluxes can be applied [m].
-
subroutine, public rgc_tracer::rgc_tracer_end(CS CS) - Parameters
cs: The control structure returned by a previous call to RGC_register_tracer.
Variables
-
integer, parameter rgc_tracer::ntr = 1 The number of tracers in this module.
-
-
namespace
rossby_front_2d_initialization¶ Initial conditions for the 2D Rossby front test.
Functions
-
subroutine, public rossby_front_2d_initialization::rossby_front_initialize_thickness(h h, G G, GV GV, param_file param_file, just_read_params just_read_params) Initialization of thicknesses in 2D Rossby front test.
- Parameters
[in] g: Grid structure[in] gv: Vertical grid structure[out] h: The thickness that is being initialized [H ~> m or kg m-2][in] param_file: A structure indicating the open file to parse for model parameter values.[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine, public rossby_front_2d_initialization::rossby_front_initialize_temperature_salinity(T T, S S, h h, G G, GV GV, param_file param_file, eqn_of_state eqn_of_state, just_read_params just_read_params) Initialization of temperature and salinity in the Rossby front test.
- Parameters
[in] g: Grid structure[in] gv: The ocean’s vertical grid structure.[out] t: Potential temperature [degC][out] s: Salinity [ppt][in] h: Thickness [H ~> m or kg m-2][in] param_file: Parameter file handleeqn_of_state: Equation of state structure[in] just_read_params: If present and true, this call will only read parameters without changing T & S.
-
subroutine, public rossby_front_2d_initialization::rossby_front_initialize_velocity(u u, v v, h h, G G, GV GV, US US, param_file param_file, just_read_params just_read_params) Initialization of u and v in the Rossby front test.
- Parameters
[in] g: Grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[out] u: i-component of velocity [m s-1][out] v: j-component of velocity [m s-1][in] h: Thickness [H ~> m or kg m-2][in] param_file: A structure indicating the open file to parse for model parameter values.[in] just_read_params: If present and true, this call will only read parameters without setting u & v.
-
real function rossby_front_2d_initialization::ypseudo(G G, lat lat) Pseudo coordinate across domain used by Hml() and dTdy() returns a coordinate from -PI/2 .. PI/2 squashed towards the center of the domain.
- Parameters
[in] g: Grid structure[in] lat: Latitude
-
real function rossby_front_2d_initialization::hml(G G, lat lat) Analytic prescription of mixed layer depth in 2d Rossby front test, in the same units as Gmax_depth.
- Parameters
[in] g: Grid structure[in] lat: Latitude
-
real function rossby_front_2d_initialization::dtdy(G G, dT dT, lat lat) Analytic prescription of mixed layer temperature gradient in 2d Rossby front test.
- Parameters
[in] g: Grid structure[in] dt: Top to bottom temperature difference[in] lat: Latitude
Variables
-
character(len=40) rossby_front_2d_initialization::mdl = "Rossby_front_2d_initialization" This module’s name.
-
real, parameter rossby_front_2d_initialization::frontfractionalwidth = 0.5 Width of front as fraction of domain [nondim].
-
real, parameter rossby_front_2d_initialization::hmlmin = 0.25 Shallowest ML as fractional depth of ocean [nondim].
-
real, parameter rossby_front_2d_initialization::hmlmax = 0.75 Deepest ML as fractional depth of ocean [nondim].
-
-
namespace
scm_cvmix_tests¶ Initial conditions and forcing for the single column model (SCM) CVMix test set.
Functions
-
subroutine, public scm_cvmix_tests::scm_cvmix_tests_ts_init(T T, S S, h h, G G, GV GV, US US, param_file param_file, just_read_params just_read_params) Initializes temperature and salinity for the SCM CVMix test example.
- Parameters
[out] t: Potential temperature [degC][out] s: Salinity [psu][in] h: Layer thickness [H ~> m or kg m-2][in] g: Grid structure[in] gv: Vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: Input parameter structure[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine, public scm_cvmix_tests::scm_cvmix_tests_surface_forcing_init(Time Time, G G, param_file param_file, CS CS) Initializes surface forcing for the CVMix test case suite.
- Parameters
[in] time: Model time[in] g: Grid structure[in] param_file: Input parameter structurecs: Parameter container
-
subroutine, public scm_cvmix_tests::scm_cvmix_tests_wind_forcing(state state, forces forces, day day, G G, US US, CS CS) - Parameters
[in] state: Surface state structure[inout] forces: A structure with the driving mechanical forces[in] day: Time in days[inout] g: Grid structure[in] us: A dimensional unit scaling typecs: Container for SCM parameters
-
subroutine, public scm_cvmix_tests::scm_cvmix_tests_buoyancy_forcing(state state, fluxes fluxes, day day, G G, CS CS) - Parameters
[in] state: Surface state structure[inout] fluxes: Surface fluxes structure[in] day: Current model time[inout] g: Grid structurecs: Container for SCM parameters
Variables
-
character(len=40) scm_cvmix_tests::mdl = "SCM_CVMix_tests" This module’s name.
-
-
namespace
seamount_initialization¶ Configures the model for the idealized seamount test case.
Functions
-
subroutine, public seamount_initialization::seamount_initialize_topography(D D, G G, param_file param_file, max_depth max_depth) Initialization of topography.
- Parameters
[in] g: The dynamic horizontal grid type[out] d: Ocean bottom depth in the units of depth_max[in] param_file: Parameter file structure[in] max_depth: Maximum ocean depth in arbitrary units
-
subroutine, public seamount_initialization::seamount_initialize_thickness(h h, G G, GV GV, US US, param_file param_file, just_read_params just_read_params) Initialization of thicknesses. This subroutine initializes the layer thicknesses to be uniform.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[out] h: The thickness that is being initialized [H ~> m or kg m-2].[in] param_file: A structure indicating the open file to parse for model parameter values.[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine, public seamount_initialization::seamount_initialize_temperature_salinity(T T, S S, h h, G G, GV GV, param_file param_file, eqn_of_state eqn_of_state, just_read_params just_read_params) Initial values for temperature and salinity.
- Parameters
[in] g: Ocean grid structure[in] gv: Vertical grid structure[out] t: Potential temperature [degC][out] s: Salinity [ppt][in] h: Layer thickness [H ~> m or kg m-2][in] param_file: Parameter file structureeqn_of_state: Equation of state structure[in] just_read_params: If present and true, this call will only read parameters without changing h.
Variables
-
character(len=40) seamount_initialization::mdl = "seamount_initialization" This module’s name.
-
-
namespace
shelfwave_initialization¶ Configures the model for the idealized shelfwave test case.
Functions
-
logical function, public shelfwave_initialization::register_shelfwave_obc(param_file param_file, CS CS, OBC_Reg OBC_Reg) Add shelfwave to OBC registry.
- Parameters
[in] param_file: parameter file.cs: shelfwave control structure.obc_reg: OBC registry.
-
subroutine, public shelfwave_initialization::shelfwave_obc_end(CS CS) Clean up the shelfwave OBC from registry.
- Parameters
cs: shelfwave control structure.
-
subroutine, public shelfwave_initialization::shelfwave_initialize_topography(D D, G G, param_file param_file, max_depth max_depth, US US) Initialization of topography.
- Parameters
[in] g: The dynamic horizontal grid type[out] d: Ocean bottom depth in m or Z if US is present[in] param_file: Parameter file structure[in] max_depth: Maximum model depth in the units of D[in] us: A dimensional unit scaling type
-
subroutine, public shelfwave_initialization::shelfwave_set_obc_data(OBC OBC, CS CS, G G, h h, Time Time) This subroutine sets the properties of flow at open boundary conditions.
- Parameters
obc: This open boundary condition type specifies whether, where, and what open boundary conditions are used.cs: tidal bay control structure.[in] g: The ocean’s grid structure.[in] h: layer thickness.[in] time: model time.
Variables
-
character(len=40) shelfwave_initialization::mdl = "shelfwave_initialization" This module’s name.
-
-
namespace
sloshing_initialization¶ Initialization for the “sloshing” internal waves configuration.
The module configures the model for the non-rotating sloshing test case.
Functions
-
subroutine, public sloshing_initialization::sloshing_initialize_topography(D D, G G, param_file param_file, max_depth max_depth) Initialization of topography.
- Parameters
[in] g: The dynamic horizontal grid type[out] d: Ocean bottom depth in the units of depth_max[in] param_file: Parameter file structure[in] max_depth: Maximum ocean depth in arbitrary units
-
subroutine, public sloshing_initialization::sloshing_initialize_thickness(h h, G G, GV GV, US US, param_file param_file, just_read_params just_read_params) Initialization of thicknesses This routine is called when THICKNESS_CONFIG is set to ‘sloshing’.
This routine initializes layer positions to set off a sloshing motion in the zonal direction in a rectangular basin. All layers have initially the same thickness but all interfaces (except bottom and sea surface) are displaced according to a half-period cosine, with maximum value on the left and minimum value on the right. This sets off a regular sloshing motion.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[out] h: The thickness that is being initialized [H ~> m or kg m-2].[in] param_file: A structure indicating the open file to parse for model parameter values.[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine, public sloshing_initialization::sloshing_initialize_temperature_salinity(T T, S S, h h, G G, GV GV, param_file param_file, eqn_of_state eqn_of_state, just_read_params just_read_params) Initialization of temperature and salinity.
This subroutine initializes linear profiles for T and S according to reference surface layer salinity and temperature and a specified range. Note that the linear distribution is set up with respect to the layer number, not the physical position).
- Parameters
[in] g: Ocean grid structure.[in] gv: The ocean’s vertical grid structure.[out] t: Potential temperature [degC].[out] s: Salinity [ppt].[in] h: Layer thickness [H ~> m or kg m-2].[in] param_file: A structure indicating the open file to parse for model parameter values.eqn_of_state: Equation of state structure.[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
-
namespace
soliton_initialization¶ Initial conditions for the Equatorial Rossby soliton test (Boyd).
Functions
-
subroutine, public soliton_initialization::soliton_initialize_thickness(h h, G G, GV GV, US US) Initialization of thicknesses in Equatorial Rossby soliton test.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[in] us: A dimensional unit scaling type[out] h: The thickness that is being initialized [H ~> m or kg m-2].
-
subroutine, public soliton_initialization::soliton_initialize_velocity(u u, v v, h h, G G) Initialization of u and v in the equatorial Rossby soliton test.
- Parameters
[in] g: Grid structure[out] u: i-component of velocity [m s-1][out] v: j-component of velocity [m s-1][in] h: Thickness [H ~> m or kg m-2]
Variables
-
character(len=40) soliton_initialization::mdl = "soliton_initialization" This module’s name.
-
-
namespace
supercritical_initialization¶ The “super critical” configuration.
The module configures the model for the “supercritical” experiment. https://marine.rutgers.edu/po/index.php?model=test-problems&title=supercritical
Functions
-
subroutine, public supercritical_initialization::supercritical_set_obc_data(OBC OBC, G G, param_file param_file) This subroutine sets the properties of flow at open boundary conditions.
- Parameters
obc: This open boundary condition type specifies whether, where, and what open boundary conditions are used.[in] g: The ocean’s grid structure.[in] param_file: Parameter file structure
-
-
namespace
tidal_bay_initialization¶ Configures the model for the “tidal_bay” experiment. tidal_bay = Tidally resonant bay from Zygmunt Kowalik’s class on tides.
Functions
-
logical function, public tidal_bay_initialization::register_tidal_bay_obc(param_file param_file, CS CS, OBC_Reg OBC_Reg) Add tidal bay to OBC registry.
- Parameters
[in] param_file: parameter file.cs: tidal bay control structure.obc_reg: OBC registry.
-
subroutine, public tidal_bay_initialization::tidal_bay_obc_end(CS CS) Clean up the tidal bay OBC from registry.
- Parameters
cs: tidal bay control structure.
-
subroutine, public tidal_bay_initialization::tidal_bay_set_obc_data(OBC OBC, CS CS, G G, h h, Time Time) This subroutine sets the properties of flow at open boundary conditions.
- Parameters
obc: This open boundary condition type specifies whether, where, and what open boundary conditions are used.cs: tidal bay control structure.[in] g: The ocean’s grid structure.[in] h: layer thickness.[in] time: model time.
-
-
namespace
tidal_forcing¶ Code by Robert Hallberg, August 2005, based on C-code by Harper Simmons, February, 2003, in turn based on code by Brian Arbic.
The main subroutine in this file calculates the total tidal contribution to the geopotential, including self-attraction and loading terms and the astronomical contributions. All options are selected with entries in a file that is parsed at run-time. Overall tides are enabled with the run-time parameter ‘TIDES=True’. Tidal constituents must be individually enabled with lines like ‘TIDE_M2=True’. This file has default values of amplitude, frequency, Love number, and phase at time 0 for the Earth’s M2, S2, N2, K2, K1, O1, P1, Q1, MF, and MM tidal constituents, but the frequency, amplitude and phase ant time 0 for each constituent can be changed at run time by setting variables like TIDE_M2_FREQ, TIDE_M2_AMP and TIDE_M2_PHASE_T0 (for M2).
In addition, the approach to calculating self-attraction and loading is set at run time. The default is to use the scalar approximation, with a coefficient TIDE_SAL_SCALAR_VALUE that must be set in the run-time file (for global runs, 0.094 is typical). Alternately, TIDAL_SAL_FROM_FILE can be set to read the SAL from a file containing the results of a previous simulation. To iterate the SAL to convergence, USE_PREVIOUS_TIDES may be useful (for details, see Arbic et al., 2004, DSR II). With TIDAL_SAL_FROM_FILE or USE_PREVIOUS_TIDES,a list of input files must be provided to describe each constituent’s properties from a previous solution.
-
namespace
user_change_diffusivity¶ Increments the diapycnal diffusivity in a specified band of latitudes and densities.
Functions
-
subroutine, public user_change_diffusivity::user_change_diff(h h, tv tv, G G, GV GV, CS CS, Kd_lay Kd_lay, Kd_int Kd_int, T_f T_f, S_f S_f, Kd_int_add Kd_int_add) This subroutine provides an interface for a user to use to modify the main code to alter the diffusivities as needed. The specific example implemented here augments the diffusivity for a specified range of latitude and coordinate potential density.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure[in] h: Layer thickness [H ~> m or kg m-2].[in] tv: A structure containing pointers to any available thermodynamic fields. Absent fields have NULL ptrs.cs: This module’s control structure.[inout] kd_lay: The diapycnal diffusivity of each layer [Z2 T-1 ~> m2 s-1].[inout] kd_int: The diapycnal diffusivity at each interface [Z2 T-1 ~> m2 s-1].[in] t_f: Temperature with massless layers filled in vertically [degC].[in] s_f: Salinity with massless layers filled in vertically [ppt].kd_int_add: The diapycnal diffusivity that is being added at each interface [Z2 T-1 ~> m2 s-1].
-
logical function user_change_diffusivity::range_ok(range range) This subroutine checks whether the 4 values of range are in ascending order.
- Return
Return value.
- Parameters
[in] range: Four values to check.
-
real function user_change_diffusivity::val_weights(val val, range range) This subroutine returns a value that goes smoothly from 0 to 1, stays at 1, and then goes smoothly back to 0 at the four values of range. The transitions are cubic, and have zero first derivatives where the curves hit 0 and 1. The values in range must be in ascending order, as can be checked by calling range_OK.
- Return
Return value.
- Parameters
[in] val: Value for which we need an answer.[in] range: Range over which the answer is non-zero.
-
subroutine, public user_change_diffusivity::user_change_diff_init(Time Time, G G, GV GV, US US, param_file param_file, diag diag, CS CS) Set up the module control structure.
- Parameters
[in] time: The current model time.[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure[in] us: A dimensional unit scaling type[in] param_file: A structure indicating the open file to parse for model parameter values.[inout] diag: A structure that is used to regulate diagnostic output.cs: A pointer that is set to point to the control structure for this module.
-
subroutine, public user_change_diffusivity::user_change_diff_end(CS CS) Clean up the module control structure.
- Parameters
cs: A pointer that is set to point to the control structure for this module.
-
-
namespace
user_initialization¶ A template of a user to code up customized initial conditions.
This subroutine initializes the fields for the simulations. The one argument passed to initialize, Time, is set to the current time of the simulation. The fields which are initialized here are:
u - Zonal velocity [m s-1].
v - Meridional velocity [m s-1].
h - Layer thickness [H ~> m or kg m-2]. (Must be positive.)
GbathyT - Basin depth [Z ~> m]. (Must be positive.)
GCoriolisBu - The Coriolis parameter [T-1 ~> s-1].
GVg_prime - The reduced gravity at each interface [L2 Z-1 T-2 ~> m s-2].
GVRlay - Layer potential density (coordinate variable) [kg m-3]. If ENABLE_THERMODYNAMICS is defined:
T - Temperature [degC].
S - Salinity [psu]. If BULKMIXEDLAYER is defined:
Rml - Mixed layer and buffer layer potential densities [kg m-3]. If SPONGE is defined:
A series of subroutine calls are made to set up the damping rates and reference profiles for all variables that are damped in the sponge.
Any user provided tracer code is also first linked through this subroutine.
These variables are all set in the set of subroutines (in this file) USER_initialize_bottom_depth, USER_initialize_thickness, USER_initialize_velocity, USER_initialize_temperature_salinity, USER_initialize_mixed_layer_density, USER_initialize_sponges, USER_set_coord, and USER_set_ref_profile.
The names of these subroutines should be self-explanatory. They start with “USER_” to indicate that they will likely have to be modified for each simulation to set the initial conditions and boundary conditions. Most of these take two arguments: an integer argument specifying whether the fields are to be calculated internally or read from a NetCDF file; and a string giving the path to that file. If the field is initialized internally, the path is ignored.
Functions
-
subroutine, public user_initialization::user_set_coord(Rlay Rlay, g_prime g_prime, GV GV, param_file param_file, eqn_of_state eqn_of_state) Set vertical coordinates.
- Parameters
[in] gv: The ocean’s vertical grid structure.[out] rlay: Layer potential density.[out] g_prime: The reduced gravity at each interface [L2 Z-1 T-2 ~> m s-2].[in] param_file: A structure indicating the open file to parse for model parameter values.eqn_of_state: Integer that selects the equation of state.
-
subroutine, public user_initialization::user_initialize_topography(D D, G G, param_file param_file, max_depth max_depth, US US) Initialize topography.
- Parameters
[in] g: The dynamic horizontal grid type[out] d: Ocean bottom depth in m or Z if US is present[in] param_file: Parameter file structure[in] max_depth: Maximum model depth in the units of D[in] us: A dimensional unit scaling type
-
subroutine, public user_initialization::user_initialize_thickness(h h, G G, GV GV, param_file param_file, just_read_params just_read_params) initialize thicknesses.
- Parameters
[in] g: The ocean’s grid structure.[in] gv: The ocean’s vertical grid structure.[out] h: The thicknesses being initialized [H ~> m or kg m-2].[in] param_file: A structure indicating the open file to parse for model parameter values.[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine, public user_initialization::user_initialize_velocity(u u, v v, G G, param_file param_file, just_read_params just_read_params) initialize velocities.
- Parameters
[in] g: Ocean grid structure.[out] u: i-component of velocity [m s-1][out] v: j-component of velocity [m/s][in] param_file: A structure indicating the open file to parse for model parameter values.[in] just_read_params: If present and true, this call will only read parameters without changing h.
-
subroutine, public user_initialization::user_init_temperature_salinity(T T, S S, G G, param_file param_file, eqn_of_state eqn_of_state, just_read_params just_read_params) This function puts the initial layer temperatures and salinities into T(:,:,:) and S(:,:,:).
- Parameters
[in] g: Ocean grid structure.[out] t: Potential temperature [degC].[out] s: Salinity [ppt].[in] param_file: A structure indicating the open file to parse for model parameter values.eqn_of_state: Integer that selects the equation of state.[in] just_read_params: If present and true, this call will only read parameters without changing T & S.
-
subroutine, public user_initialization::user_initialize_sponges(G G, GV GV, use_temp use_temp, tv tv, param_file param_file, CSp CSp, h h) Set up the sponges.
- Parameters
[in] g: Ocean grid structure.[in] gv: The ocean’s vertical grid structure.[in] use_temp: If true, temperature and salinity are state variables.[in] tv: A structure containing pointers to any available thermodynamic fields, potential temperature and salinity or mixed layer density. Absent fields have NULL ptrs.[in] param_file: A structure indicating the open file to parse for model parameter values.csp: A pointer to the sponge control structure.[in] h: Layer thicknesses [H ~> m or kg m-2].
-
subroutine, public user_initialization::user_set_obc_data(OBC OBC, tv tv, G G, param_file param_file, tr_Reg tr_Reg) This subroutine sets the properties of flow at open boundary conditions.
- Parameters
obc: This open boundary condition type specifies whether, where, and what open boundary conditions are used.[in] tv: A structure containing pointers to any available thermodynamic fields, including potential temperature and salinity or mixed layer density. Absent fields have NULL ptrs.[in] g: The ocean’s grid structure.[in] param_file: A structure indicating the open file to parse for model parameter values.tr_reg: Tracer registry.
-
subroutine, public user_initialization::user_set_rotation(G G, param_file param_file) - Parameters
[inout] g: The ocean’s grid structure[in] param_file: A structure to parse for run-time parameters
-
subroutine
write_user_log(param_file param_file)¶ Write output about the parameter values being used.
- Parameters
[in] param_file: A structure indicating the open file to parse for model parameter values.
Variables
-
logical
first_call= .true.¶ A module variable that should not be used.
-
namespace
user_revise_forcing¶ Provides a template for users to code updating the forcing fluxes.
Functions
-
subroutine, public user_revise_forcing::user_alter_forcing(state state, fluxes fluxes, day day, G G, CS CS) This subroutine sets the surface wind stresses.
- Parameters
[in] state: A structure containing fields that describe the surface state of the ocean.[inout] fluxes: A structure containing pointers to any possible forcing fields. Unused fields have NULL ptrs.[in] day: Time of the fluxes.[in] g: The ocean’s grid structure.cs: A pointer to the control structure returned by a previous call to surface_forcing_init.
-
subroutine, public user_revise_forcing::user_revise_forcing_init(param_file param_file, CS CS) Initialize the user_revise_forcing control structure.
- Parameters
[in] param_file: A structure indicating the open file to parse for model parameter values.cs: A pointer to the control structure returned by a previous call to surface_forcing_init.
Variables
-
character(len=40) user_revise_forcing::mdl = "user_revise_forcing" This module’s name.
-
-
namespace
user_shelf_init¶ This module specifies the initial values and evolving properties of the MOM6 ice shelf, using user-provided code.
Functions
-
subroutine, public user_shelf_init::user_initialize_shelf_mass(mass_shelf mass_shelf, area_shelf_h area_shelf_h, h_shelf h_shelf, hmask hmask, G G, US US, CS CS, param_file param_file, new_sim new_sim) This subroutine sets up the initial mass and area covered by the ice shelf, based on user-provided code.
- Parameters
[in] g: The ocean’s grid structure[out] mass_shelf: The ice shelf mass per unit area averaged[out] h_shelf: The ice shelf thickness [Z ~> m].[out] area_shelf_h: The area per cell covered by the ice shelf [m2].[out] hmask: A mask indicating which tracer points are[in] us: A structure containing unit conversion factorscs: A pointer to the user ice shelf control structure[in] param_file: A structure to parse for run-time parameters[in] new_sim: If true, this is a new run; otherwise it is being started from a restart file.
-
subroutine, public user_shelf_init::user_init_ice_thickness(h_shelf h_shelf, area_shelf_h area_shelf_h, hmask hmask, G G, US US, param_file param_file) This subroutine updates the ice shelf thickness, as specified by user-provided code.
- Parameters
[in] g: The ocean’s grid structure[out] h_shelf: The ice shelf thickness [m].[out] area_shelf_h: The area per cell covered by the ice shelf [m2].[out] hmask: A mask indicating which tracer points are[in] us: A structure containing unit conversion factors[in] param_file: A structure to parse for run-time parameters
-
subroutine, public user_shelf_init::user_update_shelf_mass(mass_shelf mass_shelf, area_shelf_h area_shelf_h, h_shelf h_shelf, hmask hmask, G G, CS CS, Time Time, new_sim new_sim) This subroutine updates the ice shelf mass, as specified by user-provided code.
- Parameters
[in] g: The ocean’s grid structure[inout] mass_shelf: The ice shelf mass per unit area averaged[inout] area_shelf_h: The area per cell covered by the ice shelf [m2].[inout] h_shelf: The ice shelf thickness [Z ~> m].[inout] hmask: A mask indicating which tracer points arecs: A pointer to the user ice shelf control structure[in] time: The current model time[in] new_sim: If true, this the start of a new run.
-
subroutine
write_user_log(param_file param_file)¶ This subroutine writes out the user ice shelf code version number to the model log.
- Parameters
[in] param_file: A structure to parse for run-time parameters
-
-
namespace
user_surface_forcing¶ Template for user to code up surface forcing.
Functions
-
subroutine, public user_surface_forcing::user_wind_forcing(sfc_state sfc_state, forces forces, day day, G G, US US, CS CS) This subroutine sets the surface wind stresses, forcestaux and forcestauy, in [Pa]. These are the stresses in the direction of the model grid (i.e. the same direction as the u- and v- velocities).
- Parameters
[inout] sfc_state: A structure containing fields that describe the surface state of the ocean.[inout] forces: A structure with the driving mechanical forces[in] day: The time of the fluxes[inout] g: The ocean’s grid structure[in] us: A dimensional unit scaling typecs: A pointer to the control structure returned by a previous call to user_surface_forcing_init
-
subroutine, public user_surface_forcing::user_buoyancy_forcing(sfc_state sfc_state, fluxes fluxes, day day, dt dt, G G, US US, CS CS) This subroutine specifies the current surface fluxes of buoyancy or temperature and fresh water. It may also be modified to add surface fluxes of user provided tracers.
- Parameters
[inout] sfc_state: A structure containing fields that describe the surface state of the ocean.[inout] fluxes: A structure containing thermodynamic forcing fields[in] day: The time of the fluxes[in] dt: The amount of time over which the fluxes apply [s][in] g: The ocean’s grid structure[in] us: A dimensional unit scaling typecs: A pointer to the control structure returned by a previous call to user_surface_forcing_init
-
subroutine, public user_surface_forcing::user_surface_forcing_init(Time Time, G G, US US, param_file param_file, diag diag, CS CS) This subroutine initializes the USER_surface_forcing module.
- Parameters
[in] time: The current model time[in] g: The ocean’s grid structure[in] us: A dimensional unit scaling type[in] param_file: A structure to parse for run-time parameters[in] diag: A structure that is used to regulate diagnostic output.cs: A pointer that is set to point to the control structure for this module
-
-
namespace
user_tracer_example¶ A sample tracer package that has striped initial conditions.
Original by Robert Hallberg, 2002
This file contains an example of the code that is needed to set up and use a set (in this case one) of dynamically passive tracers.
A single subroutine is called from within each file to register each of the tracers for reinitialization and advection and to register the subroutine that initializes the tracers and set up their output and the subroutine that does any tracer physics or chemistry along with diapycnal mixing (included here because some tracers may float or swim vertically or dye diapycnal processes).
Functions
-
logical function, public user_tracer_example::user_register_tracer_example(HI HI, GV GV, param_file param_file, CS CS, tr_Reg tr_Reg, restart_CS restart_CS) This subroutine is used to register tracer fields and subroutines to be used with MOM.
- Parameters
[in] hi: A horizontal index type structure[in] gv: The ocean’s vertical grid structure[in] param_file: A structure to parse for run-time parameterscs: A pointer that is set to point to the control structure for this moduletr_reg: A pointer that is set to point to the control structure for the tracer advection and diffusion modulerestart_cs: A pointer to the restart control structure
-
subroutine, public user_tracer_example::user_initialize_tracer(restart restart, day day, G G, GV GV, h h, diag diag, OBC OBC, CS CS, sponge_CSp sponge_CSp) This subroutine initializes the NTR tracer fields in tr(:,:,:,:) and it sets up the tracer output.
- Parameters
[in] restart: .true. if the fields have already been read from a restart file.[in] day: Time of the start of the run.[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][in] diag: A structure that is used to regulate diagnostic output.obc: This open boundary condition type specifies whether, where, and what open boundary conditions are used.cs: The control structure returned by a previous call to USER_register_tracer_example.sponge_csp: A pointer to the control structure for the sponges, if they are in use.
-
subroutine, public user_tracer_example::tracer_column_physics(h_old h_old, h_new h_new, ea ea, eb eb, fluxes fluxes, dt dt, G G, GV GV, CS CS) This subroutine applies diapycnal diffusion and any other column tracer physics or chemistry to the tracers from this file. This is a simple example of a set of advected passive tracers. The arguments to this subroutine are redundant in that h_new(k) = h_old(k) + ea(k) - eb(k-1) + eb(k) - ea(k+1)
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h_old: Layer thickness before entrainment [H ~> m or kg m-2].[in] h_new: Layer thickness after entrainment [H ~> m or kg m-2].[in] ea: an array to which the amount of fluid entrained[in] eb: an array to which the amount of fluid entrained[in] fluxes: A structure containing pointers to thermodynamic and tracer forcing fields. Unused fields have NULL ptrs.[in] dt: The amount of time covered by this call [s]cs: The control structure returned by a previous call to USER_register_tracer_example.
-
integer function, public user_tracer_example::user_tracer_stock(h h, stocks stocks, G G, GV GV, CS CS, names names, units units, stock_index stock_index) This function calculates the mass-weighted integral of all tracer stocks, returning the number of stocks it has calculated. If the stock_index is present, only the stock corresponding to that coded index is returned.
- Return
Return value: the number of stocks calculated here.
- Parameters
[in] g: The ocean’s grid structure[in] gv: The ocean’s vertical grid structure[in] h: Layer thicknesses [H ~> m or kg m-2][out] stocks: the mass-weighted integrated amount of each tracer, in kg times concentration units [kg conc].cs: The control structure returned by a previous call to register_USER_tracer.[out] names: The names of the stocks calculated.[out] units: The units of the stocks calculated.[in] stock_index: The coded index of a specific stock being sought.
-
subroutine, public user_tracer_example::user_tracer_surface_state(state state, h h, G G, CS CS) This subroutine extracts the surface fields from this tracer package that are to be shared with the atmosphere in coupled configurations.
- Parameters
[in] g: The ocean’s grid structure[inout] state: A structure containing fields that describe the surface state of the ocean.[in] h: Layer thicknesses [H ~> m or kg m-2]cs: The control structure returned by a previous call to register_USER_tracer.
-
subroutine, public user_tracer_example::user_tracer_example_end(CS CS) Clean up allocated memory at the end.
- Parameters
cs: The control structure returned by a previous call to register_USER_tracer.
Variables
-
integer, parameter user_tracer_example::ntr = 1 The number of tracers in this module.
-
-
file
ocean_model_MOM.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_memory.h - #include <MOM_memory_macros.h>
Compile-time memory settings.
This include file determines the compile-time memory settings. There are several variants of this file and only one should be in the search path for compilation.
Defines
-
NIGLOBAL_¶ The number of thickness grid points in the i-direction of the global domain.
-
NJGLOBAL_¶ The number of thickness grid points in the j-direction of the global domain.
-
NK_¶ The number of layers in the vertical direction.
-
NIPROC_¶ The number of processors in the i-direction.
-
NJPROC_¶ The number of processors in the j-direction.
-
MAX_FIELDS_¶ The maximum permitted number (each) of restart variables, time derivatives, etc.
This is mostly used for the size of pointer arrays, so it should be set generously.
-
NIHALO_¶ The number of memory halo cells on each side of the computational domain in the i-direction.
-
NJHALO_¶ The number of memory halo cells on each side of the computational domain in the j-direction.
-
SYMMETRIC_MEMORY_¶ If SYMMETRIC_MEMORY_() is defined, the velocity point data domain includes every face of the thickness points.
In other words, some arrays are larger than others, depending on where they are on the staggered grid.
-
-
file
atmos_ocean_fluxes.F90
-
file
MESO_surface_forcing.F90 - #include “version_variable.h”
-
file
MOM_driver.F90 - #include <MOM_memory.h>#include “version_variable.h”
Functions
-
program
mom_main()¶
-
program
-
file
MOM_surface_forcing.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
Neverland_surface_forcing.F90
-
file
user_surface_forcing.F90
-
file
coord_adapt.F90 - #include <MOM_memory.h>
-
file
coord_hycom.F90
-
file
coord_rho.F90
-
file
coord_sigma.F90
-
file
coord_slight.F90
-
file
coord_zlike.F90
-
file
MOM_ALE.F90 - #include <MOM_memory.h>
-
file
MOM_regridding.F90 - #include <MOM_memory.h>
-
file
MOM_remapping.F90 - #include <MOM_memory.h>
-
file
P1M_functions.F90
-
file
P3M_functions.F90
-
file
PCM_functions.F90
-
file
PLM_functions.F90
-
file
polynomial_functions.F90
-
file
PPM_functions.F90
-
file
PQM_functions.F90
-
file
regrid_consts.F90
-
file
regrid_edge_slopes.F90
-
file
regrid_edge_values.F90
-
file
regrid_interp.F90
-
file
regrid_solvers.F90
-
file
MOM.F90 - #include <MOM_memory.h>
-
file
MOM_barotropic.F90 - #include <MOM_memory.h>
-
file
MOM_boundary_update.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_checksum_packages.F90 - #include <MOM_memory.h>
-
file
MOM_continuity.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_continuity_PPM.F90 - #include <MOM_memory.h>
-
file
MOM_CoriolisAdv.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_dynamics_split_RK2.F90 - #include <MOM_memory.h>
-
file
MOM_dynamics_unsplit.F90 - #include <MOM_memory.h>
-
file
MOM_dynamics_unsplit_RK2.F90 - #include <MOM_memory.h>
-
file
MOM_forcing_type.F90 - #include <MOM_memory.h>
-
file
MOM_grid.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_interface_heights.F90 - #include <MOM_memory.h>
-
file
MOM_isopycnal_slopes.F90 - #include <MOM_memory.h>
-
file
MOM_open_boundary.F90 - #include <MOM_memory.h>
Functions
-
integer function parse_segment_str::interpret_int_expr(string string, imax imax) - Parameters
[in] string: Integer in form or I, N or N-I[in] imax: Value to replace ‘N’ with
-
-
file
MOM_PressureForce.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_PressureForce_analytic_FV.F90 - #include <MOM_memory.h>
-
file
MOM_PressureForce_blocked_AFV.F90 - #include <MOM_memory.h>
-
file
MOM_PressureForce_Montgomery.F90 - #include <MOM_memory.h>
-
file
MOM_transcribe_grid.F90
-
file
MOM_unit_tests.F90
-
file
MOM_variables.F90 - #include <MOM_memory.h>
-
file
MOM_verticalGrid.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_debugging.F90 - #include “version_variable.h”
-
file
MOM_diagnostics.F90 - #include <MOM_memory.h>
-
file
MOM_obsolete_diagnostics.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_obsolete_params.F90 - #include <MOM_memory.h>
-
file
MOM_PointAccel.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_sum_output.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_wave_speed.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_wave_structure.F90 - #include <MOM_memory.h>
-
file
MOM_EOS.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_EOS_linear.F90 - #include <MOM_memory.h>
-
file
MOM_EOS_NEMO.F90
-
file
MOM_EOS_TEOS10.F90
-
file
MOM_EOS_UNESCO.F90
-
file
MOM_EOS_Wright.F90 - #include <MOM_memory.h>
-
file
MOM_TFreeze.F90
-
file
_Diagnostics.dox
-
file
_Horizontal_indexing.dox
-
file
_Runtime_parameter_system.dox
-
file
MOM_checksums.F90 - #include “version_variable.h”
Functions
-
integer function zchksum::subchk(array array, scale scale) - Parameters
[in] array: The array to be checksummed[in] scale: A scaling factor for this array.
-
subroutine
substats(array array, aMean aMean, aMin aMin, aMax aMax)¶ - Parameters
[in] array: The array to be checksummed
-
integer function chksum_h_2d::subchk(array array, HI HI, di di, dj dj, scale scale) - Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] di: i- direction array shift for this checksum[in] dj: j- direction array shift for this checksum[in] scale: A scaling factor for this array.
-
subroutine
substats(HI HI, array array, aMean aMean, aMin aMin, aMax aMax)¶ - Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed
-
integer function chksum_b_2d::subchk(array array, HI HI, di di, dj dj, scale scale) - Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] di: i- direction array shift for this checksum[in] dj: j- direction array shift for this checksum[in] scale: A scaling factor for this array.
-
subroutine
substats(HI HI, array array, sym_stats sym_stats, aMean aMean, aMin aMin, aMax aMax)¶ - Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] sym_stats: If true, evaluate the statistics on the full symmetric computational domain.
-
integer function chksum_u_2d::subchk(array array, HI HI, di di, dj dj, scale scale) - Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] di: i- direction array shift for this checksum[in] dj: j- direction array shift for this checksum[in] scale: A scaling factor for this array.
-
subroutine
substats(HI HI, array array, sym_stats sym_stats, aMean aMean, aMin aMin, aMax aMax)¶ - Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] sym_stats: If true, evaluate the statistics on the full symmetric computational domain.
-
integer function chksum_v_2d::subchk(array array, HI HI, di di, dj dj, scale scale) - Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] di: i- direction array shift for this checksum[in] dj: j- direction array shift for this checksum[in] scale: A scaling factor for this array.
-
subroutine
substats(HI HI, array array, sym_stats sym_stats, aMean aMean, aMin aMin, aMax aMax)¶ - Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] sym_stats: If true, evaluate the statistics on the full symmetric computational domain.
-
integer function chksum_h_3d::subchk(array array, HI HI, di di, dj dj, scale scale) - Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] di: i- direction array shift for this checksum[in] dj: j- direction array shift for this checksum[in] scale: A scaling factor for this array.
-
subroutine
substats(HI HI, array array, aMean aMean, aMin aMin, aMax aMax)¶ - Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed
-
integer function chksum_b_3d::subchk(array array, HI HI, di di, dj dj, scale scale) - Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] di: i- direction array shift for this checksum[in] dj: j- direction array shift for this checksum[in] scale: A scaling factor for this array.
-
subroutine
substats(HI HI, array array, sym_stats sym_stats, aMean aMean, aMin aMin, aMax aMax)¶ - Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] sym_stats: If true, evaluate the statistics on the full symmetric computational domain.
-
integer function chksum_u_3d::subchk(array array, HI HI, di di, dj dj, scale scale) - Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] di: i- direction array shift for this checksum[in] dj: j- direction array shift for this checksum[in] scale: A scaling factor for this array.
-
subroutine
substats(HI HI, array array, sym_stats sym_stats, aMean aMean, aMin aMin, aMax aMax)¶ - Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] sym_stats: If true, evaluate the statistics on the full symmetric computational domain.
-
integer function chksum_v_3d::subchk(array array, HI HI, di di, dj dj, scale scale) - Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] di: i- direction array shift for this checksum[in] dj: j- direction array shift for this checksum[in] scale: A scaling factor for this array.
-
subroutine
substats(HI HI, array array, sym_stats sym_stats, aMean aMean, aMin aMin, aMax aMax)¶ - Parameters
[in] hi: A horizontal index type[in] array: The array to be checksummed[in] sym_stats: If true, evaluate the statistics on the full symmetric computational domain.[out] amax: Mean/min/max of array over domain
-
-
file
MOM_coms.F90
-
file
MOM_constants.F90
-
file
MOM_cpu_clock.F90
-
file
MOM_diag_manager_wrapper.F90
-
file
MOM_diag_mediator.F90
-
file
MOM_diag_remap.F90
-
file
MOM_diag_vkernels.F90
-
file
MOM_document.F90
-
file
MOM_domains.F90
-
file
MOM_dyn_horgrid.F90
-
file
MOM_error_handler.F90
-
file
MOM_file_parser.F90
-
file
MOM_get_input.F90
-
file
MOM_hor_index.F90 - #include “version_variable.h”
-
file
MOM_horizontal_regridding.F90 - #include <MOM_memory.h>
-
file
MOM_intrinsic_functions.F90
-
file
MOM_io.F90 - #include “version_variable.h”
-
file
MOM_memory_macros.h Memory macros.
This is a header file to define macros for static and dynamic memory allocation. Define STATIC_MEMORY_ in MOM_memory.h for static memory allocation. Otherwise dynamic memory allocation will be assumed. For explanation of symmetric and non-symmetric memory modes see Horizontal indexing and memory.
Defines
-
DEALLOC_(x)¶ Deallocates array x when using dynamic memory mode. Does nothing in static memory mode.
-
ALLOC_(x)¶ Allocates array x when using dynamic memory mode. Does nothing in static memory mode.
-
ALLOCABLE_¶ Attaches the ALLOCATABLE attribute to an array in dynamic memory mode. Does nothing in static memory mode.
-
PTR_¶ Attaches the POINTER attribute to an array in dynamic memory mode. Does nothing in static memory mode.
-
TO_NULL_¶ Nullify a pointer in dynamic memory mode. Does nothing in static memory mode.
-
NIMEM_¶ Expands to : in dynamic memory mode, or is the i-shape of a tile in static memory mode.
Use for heap (ALLOCABLE_ or PTR_) variables at h- or v- points.
-
NJMEM_¶ Expands to : in dynamic memory mode, or is the j-shape of a tile in static memory mode.
Use for heap (ALLOCABLE_ or PTR_) variables at h- or u- points.
-
NIMEMB_PTR_¶ Expands to : in dynamic memory mode, or to NIMEMB_ in static memory mode.
Use for heap (ALLOCABLE_ or PTR_) variables at h- or v- points.
-
NJMEMB_PTR_¶ Expands to : in dynamic memory mode, or to NJMEMB_ in static memory mode.
Use for heap (ALLOCABLE_ or PTR_) variables at h- or u- points.
-
NIMEMB_¶ Expands to : or 0: in dynamic memory mode, or is the staggered i-shape of a tile in static memory mode.
Use for heap (ALLOCABLE_ or PTR_) variables at q- or u- points.
-
NJMEMB_¶ Expands to : or 0: in dynamic memory mode, or is the staggered j-shape of a tile in static memory mode.
Use for heap (ALLOCABLE_ or PTR_) variables at q- or v- points.
-
NIMEMB_SYM_¶ Expands to 0: in dynamic memory mode, or is the staggered i-shape of a tile in static memory mode.
Use for always-symmetric heap (ALLOCABLE_ or PTR_) variables at q- or u- points.
-
NJMEMB_SYM_¶ Expands to 0: in dynamic memory mode, or is the staggered j-shape of a tile in static memory mode.
Use for always-symmetric heap (ALLOCABLE_ or PTR_) variables at q- or v- points.
-
NKMEM_¶ Expands to : in dynamic memory mode or is to the number of layers in static memory mode.
Use for heap (ALLOCABLE_ or PTR_) layer variables.
-
NKMEM0_¶ Expands to 0: in dynamic memory mode or to 0:NK_ in static memory mode.
Use for heap (ALLOCABLE_ or PTR_) interface variables.
-
NK_INTERFACE_¶ Expands to : in dynamic memory mode or to NK_+1 in static memory mode.
Use for heap (ALLOCABLE_ or PTR_) interface variables.
-
C1_¶ Expands to : or 1. UNKNOWN PURPOSE!
-
C2_¶ Expands to : or 2. UNKNOWN PURPOSE!
-
C3_¶ Expands to : or 3. UNKNOWN PURPOSE!
-
SZI_(G)¶ The i-shape of a dummy argument staggered at h- or v-points.
-
SZJ_(G)¶ The j-shape of a dummy argument staggered at h- or u-points.
-
SZK_(G)¶ The k-shape of a layer dummy argument.
-
SZK0_(G)¶ The k-shape of an interface dummy argument.
-
SZIB_(G)¶ The i-shape of a dummy argument staggered at q- or u-points.
-
SZJB_(G)¶ The j-shape of a dummy argument staggered at q- or v-points.
-
SZIBS_(G)¶ The i-shape of a symmetric dummy argument staggered at q- or u-points.
-
SZJBS_(G)¶ The j-shape of a symmetric dummy argument staggered at q- or v-points.
-
SZDI_(G)¶ The i-shape of a dynamic dummy argument staggered at h- or v-points.
-
SZDIB_(G)¶ The i-shape of a dynamic dummy argument staggered at q- or u-points.
-
SZDJ_(G)¶ The j-shape of a dynamic dummy argument staggered at h- or u-points.
-
SZDJB_(G)¶ The j-shape of a dynamic dummy argument staggered at q- or v-points.
-
-
file
MOM_restart.F90
-
file
MOM_safe_alloc.F90
-
file
MOM_spatial_means.F90 - #include <MOM_memory.h>
-
file
MOM_string_functions.F90
-
file
MOM_time_manager.F90
-
file
MOM_unit_scaling.F90 - #include “version_variable.h”
-
file
MOM_write_cputime.F90 - #include “version_variable.h”
-
file
version_variable.h Functions
-
character(len = *)¶
-
-
file
MOM_ice_shelf.F90 - #include <MOM_memory.h>
Defines
-
GRID_SYM_¶
-
-
file
MOM_ice_shelf_dynamics.F90 - #include <MOM_memory.h>
-
file
MOM_ice_shelf_initialize.F90 - #include <MOM_memory.h>
-
file
MOM_ice_shelf_state.F90
-
file
MOM_marine_ice.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
user_shelf_init.F90 - #include <MOM_memory.h>
-
file
midas_vertmap.F90
-
file
MOM_coord_initialization.F90
-
file
MOM_fixed_initialization.F90
-
file
MOM_grid_initialize.F90
-
file
MOM_shared_initialization.F90
-
file
MOM_state_initialization.F90 - #include <MOM_memory.h>#include “version_variable.h”
Functions
-
real function initialize_velocity_circular::my_psi(ig ig, jg jg) Returns the value of a circular stream function at (ig,jg)
- Parameters
ig: Global i-indexjg: Global j-index
-
-
file
MOM_tracer_initialization_from_Z.F90 - #include <MOM_memory.h>
-
file
MOM_oda_driver.F90 - #include <MOM_memory.h>
-
file
MOM_hor_visc.F90 - #include <MOM_memory.h>
-
file
MOM_internal_tides.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_lateral_mixing_coeffs.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_MEKE.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_MEKE_types.F90
-
file
MOM_mixed_layer_restrat.F90 - #include <MOM_memory.h>
-
file
MOM_thickness_diffuse.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_tidal_forcing.F90 - #include <MOM_memory.h>
-
file
MOM_ALE_sponge.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_bkgnd_mixing.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_bulk_mixed_layer.F90 - #include <MOM_memory.h>
-
file
MOM_CVMix_conv.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_CVMix_ddiff.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_CVMix_KPP.F90 - #include “MOM_memory.h”#include “version_variable.h”
Defines
-
__DO_SAFETY_CHECKS__¶
-
-
file
MOM_CVMix_shear.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_diabatic_aux.F90 - #include <MOM_memory.h>#include “version_variable.h”
Defines
-
_OLD_ALG_¶
-
-
file
MOM_diabatic_driver.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_diapyc_energy_req.F90 - #include “version_variable.h”
-
file
MOM_energetic_PBL.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_entrain_diffusive.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_full_convection.F90 - #include <MOM_memory.h>
-
file
MOM_geothermal.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_internal_tide_input.F90 - #include <MOM_memory.h>
-
file
MOM_kappa_shear.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_opacity.F90 - #include <MOM_memory.h>
-
file
MOM_regularize_layers.F90 - #include <MOM_memory.h>
-
file
MOM_set_diffusivity.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_set_viscosity.F90 - #include <MOM_memory.h>
-
file
MOM_sponge.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_tidal_mixing.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_vert_friction.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
_Advection.dox
-
file
advection_test_tracer.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
boundary_impulse_tracer.F90
-
file
DOME_tracer.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
dye_example.F90 - #include <MOM_memory.h>
-
file
dyed_obc_tracer.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
ideal_age_example.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
ISOMIP_tracer.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_generic_tracer.F90 - #include <MOM_memory.h>
-
file
MOM_neutral_diffusion.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_neutral_diffusion_aux.F90
-
file
MOM_OCMIP2_CFC.F90 - #include <MOM_memory.h>
-
file
MOM_offline_aux.F90 - #include “MOM_memory.h”#include “version_variable.h”
-
file
MOM_offline_main.F90 - #include “MOM_memory.h”#include “version_variable.h”
-
file
MOM_tracer_advect.F90 - #include <MOM_memory.h>
-
file
MOM_tracer_diabatic.F90 - #include <MOM_memory.h>
-
file
MOM_tracer_flow_control.F90 - #include <MOM_memory.h>
-
file
MOM_tracer_hor_diff.F90 - #include <MOM_memory.h>
-
file
MOM_tracer_registry.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_tracer_Z_init.F90 - #include <MOM_memory.h>
-
file
oil_tracer.F90 - #include <MOM_memory.h>
-
file
pseudo_salt_tracer.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
RGC_tracer.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
tracer_example.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
adjustment_initialization.F90
-
file
baroclinic_zone_initialization.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
benchmark_initialization.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
BFB_initialization.F90 - #include <MOM_memory.h>
-
file
BFB_surface_forcing.F90 - #include “version_variable.h”
-
file
circle_obcs_initialization.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
dense_water_initialization.F90 - #include <MOM_memory.h>
-
file
DOME2d_initialization.F90 - #include <MOM_memory.h>
-
file
DOME_initialization.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
dumbbell_initialization.F90 - #include <MOM_memory.h>
-
file
dumbbell_surface_forcing.F90
-
file
dyed_channel_initialization.F90 - #include <MOM_memory.h>
-
file
dyed_obcs_initialization.F90 - #include <MOM_memory.h>
-
file
external_gwave_initialization.F90 - #include <MOM_memory.h>
-
file
Idealized_Hurricane.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
ISOMIP_initialization.F90 - #include <MOM_memory.h>
-
file
Kelvin_initialization.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
lock_exchange_initialization.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
MOM_controlled_forcing.F90 - #include <MOM_memory.h>
-
file
MOM_wave_interface.F90 - #include <MOM_memory.h>
-
file
Neverland_initialization.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
Phillips_initialization.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
RGC_initialization.F90 - #include <MOM_memory.h>
-
file
Rossby_front_2d_initialization.F90 - #include <MOM_memory.h>
-
file
SCM_CVMix_tests.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
seamount_initialization.F90 - #include <MOM_memory.h>
-
file
shelfwave_initialization.F90 - #include <MOM_memory.h>
-
file
sloshing_initialization.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
soliton_initialization.F90 - #include <MOM_memory.h>
-
file
supercritical_initialization.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
tidal_bay_initialization.F90
-
file
user_change_diffusivity.F90 - #include <MOM_memory.h>
-
file
user_initialization.F90 - #include <MOM_memory.h>#include “version_variable.h”
-
file
user_revise_forcing.F90 - #include “version_variable.h”
-
page
Diagnostics Controlling run-time diagnostics and how to add diagnostics to the code.
Controlling run-time diagnostics and how to add diagnostics to the code
MOM6 diagnostics are orchestrated via the FMS diag_manager, as for previous versions of MOM. However, because MOM6 is a general coordinate model, the model native-coordinae output can be less familiar to users of earlier generations of MOM. To alleviate this problem, MOM6 provides both “native” and “remapped” diagnostics; the former being diagnostics in the actual model coordinate space, and the latter in user-defined coordinates.
-
page
Horizontal_indexing Conventions for staggering of variables and loops over 2d/3d arrays.
Conventions for staggering of variables and loops over 2d/3d arrays
MOM6 is written in Fortran90 and uses the
i,j,korder of indexing.icorresponds to the fastest index (stride-1 in memory) and thus should be the inner-most loop variable. We often refer to the i-direction as the x- or zonal direction, and similarly to the j-direction as y- or meridional direction. The model can use curvilinear grids/coordinates in the horizontal and so these labels have loose meanings but convenient.
-
page
Runtime_parameter_system How run-time parameters work in MOM6.
How run-time parameters work in MOM6
MOM6 has an extensive set of parameters that are set at run-time by parsing an input file. Many parameters have default values and are not required to be in the input file, although there are a number of parameters that must be set for the model to run. The numerous examples provided with the MOM6 code mostly differ in their run-time parameters (although some add other components, like sea-ice), and comparison between these examples is an excellent way to get a broad overview of many of MOM6’s parameters and how they might be set.
-
page
Advection Tracer transport schemes.
Tracer transport schemes
MOM6 implements a generalised tracer advection scheme, which is a combination of the modified flux advection scheme (Easter, 1993) with reconstructed tracer distributions. The tracer distributions may be piecewise linear (PLM) or piecewise parabolic (PPM), which may itself use either the Colella and Woodward (CW84) or Huynh (H3) reconstruction.
-
dir
/home/docs/checkouts/readthedocs.org/user_builds/mom6-doctesting/checkouts/breathe/src/ALE
-
dir
/home/docs/checkouts/readthedocs.org/user_builds/mom6-doctesting/checkouts/breathe/config_src
-
dir
/home/docs/checkouts/readthedocs.org/user_builds/mom6-doctesting/checkouts/breathe/src/core
-
dir
/home/docs/checkouts/readthedocs.org/user_builds/mom6-doctesting/checkouts/breathe/config_src/coupled_driver
-
dir
/home/docs/checkouts/readthedocs.org/user_builds/mom6-doctesting/checkouts/breathe/src/diagnostics
-
dir
/home/docs/checkouts/readthedocs.org/user_builds/mom6-doctesting/checkouts/breathe/config_src/dynamic_symmetric
-
dir
/home/docs/checkouts/readthedocs.org/user_builds/mom6-doctesting/checkouts/breathe/src/equation_of_state
-
dir
/home/docs/checkouts/readthedocs.org/user_builds/mom6-doctesting/checkouts/breathe/src/framework
-
dir
/home/docs/checkouts/readthedocs.org/user_builds/mom6-doctesting/checkouts/breathe/src/ice_shelf
-
dir
/home/docs/checkouts/readthedocs.org/user_builds/mom6-doctesting/checkouts/breathe/src/initialization
-
dir
/home/docs/checkouts/readthedocs.org/user_builds/mom6-doctesting/checkouts/breathe/src/parameterizations/lateral
-
dir
/home/docs/checkouts/readthedocs.org/user_builds/mom6-doctesting/checkouts/breathe/src/ocean_data_assim
-
dir
/home/docs/checkouts/readthedocs.org/user_builds/mom6-doctesting/checkouts/breathe/src/parameterizations
-
dir
/home/docs/checkouts/readthedocs.org/user_builds/mom6-doctesting/checkouts/breathe/config_src/solo_driver
-
dir
/home/docs/checkouts/readthedocs.org/user_builds/mom6-doctesting/checkouts/breathe/src
-
dir
/home/docs/checkouts/readthedocs.org/user_builds/mom6-doctesting/checkouts/breathe/src/tracer
-
dir
/home/docs/checkouts/readthedocs.org/user_builds/mom6-doctesting/checkouts/breathe/src/user
-
dir
/home/docs/checkouts/readthedocs.org/user_builds/mom6-doctesting/checkouts/breathe/src/parameterizations/vertical